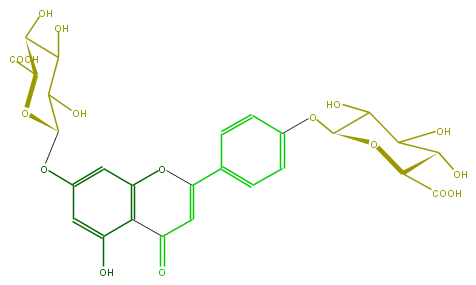
Mol:FL3FAAGS0027
From Metabolomics.JP
(Difference between revisions)
| Line 1: | Line 1: | ||
| − | + | ||
| − | + | ||
| − | Copyright: ARM project http://www.metabolome.jp/ | + | Copyright: ARM project http://www.metabolome.jp/ |
| − | 44 48 0 0 0 0 0 0 0 0999 V2000 | + | 44 48 0 0 0 0 0 0 0 0999 V2000 |
| − | -3.5550 -1.0308 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.5550 -1.0308 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.5550 -1.8403 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.5550 -1.8403 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.8539 -2.2451 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.8539 -2.2451 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.1527 -1.8403 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.1527 -1.8403 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.1527 -1.0308 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.1527 -1.0308 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.8539 -0.6260 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.8539 -0.6260 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.4516 -2.2451 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.4516 -2.2451 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.7505 -1.8403 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.7505 -1.8403 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.7505 -1.0308 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.7505 -1.0308 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.4516 -0.6260 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.4516 -0.6260 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.4516 -3.0606 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.4516 -3.0606 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.0496 -0.6261 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.0496 -0.6261 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.6649 -1.0386 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.6649 -1.0386 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.3795 -0.6261 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.3795 -0.6261 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.3795 0.1990 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.3795 0.1990 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.6649 0.6115 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.6649 0.6115 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.0496 0.1990 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.0496 0.1990 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.2559 -0.6261 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.2559 -0.6261 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.8539 -3.0544 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.8539 -3.0544 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.0938 0.6114 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.0938 0.6114 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.4345 0.6875 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.4345 0.6875 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.5884 0.1989 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.5884 0.1989 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.5362 -0.0382 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.5362 -0.0382 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.2205 -0.7434 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.2205 -0.7434 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 5.0668 -0.2549 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 5.0668 -0.2549 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.1189 -0.0176 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.1189 -0.0176 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.6729 0.9086 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.6729 0.9086 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 5.0936 0.2652 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 5.0936 0.2652 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 5.4959 -0.7550 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 5.4959 -0.7550 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.6871 0.6903 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.6871 0.6903 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.8953 0.2331 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.8953 0.2331 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.1466 1.1121 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.1466 1.1121 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.8953 1.9966 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.8953 1.9966 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.6871 2.4538 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.6871 2.4538 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.4360 1.5747 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.4360 1.5747 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.2837 3.0606 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.2837 3.0606 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.9145 2.7100 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.9145 2.7100 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.4849 0.6828 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.4849 0.6828 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.9963 1.9720 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.9963 1.9720 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.9963 3.0117 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.9963 3.0117 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -5.8968 1.4521 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -5.8968 1.4521 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.9963 -1.1913 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.9963 -1.1913 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 5.8968 -0.6714 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 5.8968 -0.6714 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.9963 -2.2311 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.9963 -2.2311 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1 2 1 0 0 0 0 | + | 1 2 1 0 0 0 0 |
| − | 2 3 2 0 0 0 0 | + | 2 3 2 0 0 0 0 |
| − | 3 4 1 0 0 0 0 | + | 3 4 1 0 0 0 0 |
| − | 4 5 2 0 0 0 0 | + | 4 5 2 0 0 0 0 |
| − | 5 6 1 0 0 0 0 | + | 5 6 1 0 0 0 0 |
| − | 6 1 2 0 0 0 0 | + | 6 1 2 0 0 0 0 |
| − | 4 7 1 0 0 0 0 | + | 4 7 1 0 0 0 0 |
| − | 7 8 1 0 0 0 0 | + | 7 8 1 0 0 0 0 |
| − | 8 9 2 0 0 0 0 | + | 8 9 2 0 0 0 0 |
| − | 9 10 1 0 0 0 0 | + | 9 10 1 0 0 0 0 |
| − | 10 5 1 0 0 0 0 | + | 10 5 1 0 0 0 0 |
| − | 7 11 2 0 0 0 0 | + | 7 11 2 0 0 0 0 |
| − | 9 12 1 0 0 0 0 | + | 9 12 1 0 0 0 0 |
| − | 12 13 2 0 0 0 0 | + | 12 13 2 0 0 0 0 |
| − | 13 14 1 0 0 0 0 | + | 13 14 1 0 0 0 0 |
| − | 14 15 2 0 0 0 0 | + | 14 15 2 0 0 0 0 |
| − | 15 16 1 0 0 0 0 | + | 15 16 1 0 0 0 0 |
| − | 16 17 2 0 0 0 0 | + | 16 17 2 0 0 0 0 |
| − | 17 12 1 0 0 0 0 | + | 17 12 1 0 0 0 0 |
| − | 1 18 1 0 0 0 0 | + | 1 18 1 0 0 0 0 |
| − | 3 19 1 0 0 0 0 | + | 3 19 1 0 0 0 0 |
| − | 15 20 1 0 0 0 0 | + | 15 20 1 0 0 0 0 |
| − | 21 22 1 0 0 0 0 | + | 21 22 1 0 0 0 0 |
| − | 22 23 1 1 0 0 0 | + | 22 23 1 1 0 0 0 |
| − | 25 24 1 1 0 0 0 | + | 25 24 1 1 0 0 0 |
| − | 25 26 1 0 0 0 0 | + | 25 26 1 0 0 0 0 |
| − | 26 21 1 0 0 0 0 | + | 26 21 1 0 0 0 0 |
| − | 21 27 1 0 0 0 0 | + | 21 27 1 0 0 0 0 |
| − | 26 28 1 0 0 0 0 | + | 26 28 1 0 0 0 0 |
| − | 25 29 1 0 0 0 0 | + | 25 29 1 0 0 0 0 |
| − | 22 20 1 0 0 0 0 | + | 22 20 1 0 0 0 0 |
| − | 23 24 1 1 0 0 0 | + | 23 24 1 1 0 0 0 |
| − | 31 30 1 1 0 0 0 | + | 31 30 1 1 0 0 0 |
| − | 31 32 1 0 0 0 0 | + | 31 32 1 0 0 0 0 |
| − | 32 33 1 0 0 0 0 | + | 32 33 1 0 0 0 0 |
| − | 33 34 1 0 0 0 0 | + | 33 34 1 0 0 0 0 |
| − | 34 35 1 1 0 0 0 | + | 34 35 1 1 0 0 0 |
| − | 35 30 1 1 0 0 0 | + | 35 30 1 1 0 0 0 |
| − | 34 36 1 0 0 0 0 | + | 34 36 1 0 0 0 0 |
| − | 33 37 1 0 0 0 0 | + | 33 37 1 0 0 0 0 |
| − | 18 31 1 0 0 0 0 | + | 18 31 1 0 0 0 0 |
| − | 32 38 1 0 0 0 0 | + | 32 38 1 0 0 0 0 |
| − | 39 40 2 0 0 0 0 | + | 39 40 2 0 0 0 0 |
| − | 39 41 1 0 0 0 0 | + | 39 41 1 0 0 0 0 |
| − | 35 39 1 0 0 0 0 | + | 35 39 1 0 0 0 0 |
| − | 42 43 2 0 0 0 0 | + | 42 43 2 0 0 0 0 |
| − | 42 44 1 0 0 0 0 | + | 42 44 1 0 0 0 0 |
| − | 24 42 1 0 0 0 0 | + | 24 42 1 0 0 0 0 |
| − | M STY 1 1 SUP | + | M STY 1 1 SUP |
| − | M SLB 1 1 1 | + | M SLB 1 1 1 |
| − | M SAL 1 3 39 40 41 | + | M SAL 1 3 39 40 41 |
| − | M SBL 1 1 45 | + | M SBL 1 1 45 |
| − | M SMT 1 ^COOH | + | M SMT 1 ^COOH |
| − | M SBV 1 45 0.5604 -0.3973 | + | M SBV 1 45 0.5604 -0.3973 |
| − | M STY 1 2 SUP | + | M STY 1 2 SUP |
| − | M SLB 1 2 2 | + | M SLB 1 2 2 |
| − | M SAL 2 3 42 43 44 | + | M SAL 2 3 42 43 44 |
| − | M SBL 2 1 48 | + | M SBL 2 1 48 |
| − | M SMT 2 COOH | + | M SMT 2 COOH |
| − | M SBV 2 48 -0.7758 0.4479 | + | M SBV 2 48 -0.7758 0.4479 |
| − | S SKP 5 | + | S SKP 5 |
| − | ID FL3FAAGS0027 | + | ID FL3FAAGS0027 |
| − | FORMULA C27H26O17 | + | FORMULA C27H26O17 |
| − | EXACTMASS 622.116999406 | + | EXACTMASS 622.116999406 |
| − | AVERAGEMASS 622.48514 | + | AVERAGEMASS 622.48514 |
| − | SMILES C(Oc(c2)ccc(C(=C5)Oc(c3)c(C5=O)c(cc3OC(O4)C(C(C(O)C4C(O)=O)O)O)O)c2)(O1)C(C(C(O)C1C(O)=O)O)O | + | SMILES C(Oc(c2)ccc(C(=C5)Oc(c3)c(C5=O)c(cc3OC(O4)C(C(C(O)C4C(O)=O)O)O)O)c2)(O1)C(C(C(O)C1C(O)=O)O)O |
M END | M END | ||
| − | |||
Latest revision as of 09:00, 14 March 2009
Copyright: ARM project http://www.metabolome.jp/
44 48 0 0 0 0 0 0 0 0999 V2000
-3.5550 -1.0308 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.5550 -1.8403 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.8539 -2.2451 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.1527 -1.8403 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.1527 -1.0308 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.8539 -0.6260 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.4516 -2.2451 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.7505 -1.8403 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.7505 -1.0308 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.4516 -0.6260 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.4516 -3.0606 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.0496 -0.6261 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.6649 -1.0386 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.3795 -0.6261 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.3795 0.1990 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.6649 0.6115 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.0496 0.1990 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.2559 -0.6261 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.8539 -3.0544 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.0938 0.6114 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.4345 0.6875 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.5884 0.1989 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.5362 -0.0382 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
4.2205 -0.7434 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
5.0668 -0.2549 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.1189 -0.0176 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.6729 0.9086 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
5.0936 0.2652 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
5.4959 -0.7550 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.6871 0.6903 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.8953 0.2331 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.1466 1.1121 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.8953 1.9966 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.6871 2.4538 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.4360 1.5747 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.2837 3.0606 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.9145 2.7100 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.4849 0.6828 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.9963 1.9720 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.9963 3.0117 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-5.8968 1.4521 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
4.9963 -1.1913 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
5.8968 -0.6714 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
4.9963 -2.2311 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1 2 1 0 0 0 0
2 3 2 0 0 0 0
3 4 1 0 0 0 0
4 5 2 0 0 0 0
5 6 1 0 0 0 0
6 1 2 0 0 0 0
4 7 1 0 0 0 0
7 8 1 0 0 0 0
8 9 2 0 0 0 0
9 10 1 0 0 0 0
10 5 1 0 0 0 0
7 11 2 0 0 0 0
9 12 1 0 0 0 0
12 13 2 0 0 0 0
13 14 1 0 0 0 0
14 15 2 0 0 0 0
15 16 1 0 0 0 0
16 17 2 0 0 0 0
17 12 1 0 0 0 0
1 18 1 0 0 0 0
3 19 1 0 0 0 0
15 20 1 0 0 0 0
21 22 1 0 0 0 0
22 23 1 1 0 0 0
25 24 1 1 0 0 0
25 26 1 0 0 0 0
26 21 1 0 0 0 0
21 27 1 0 0 0 0
26 28 1 0 0 0 0
25 29 1 0 0 0 0
22 20 1 0 0 0 0
23 24 1 1 0 0 0
31 30 1 1 0 0 0
31 32 1 0 0 0 0
32 33 1 0 0 0 0
33 34 1 0 0 0 0
34 35 1 1 0 0 0
35 30 1 1 0 0 0
34 36 1 0 0 0 0
33 37 1 0 0 0 0
18 31 1 0 0 0 0
32 38 1 0 0 0 0
39 40 2 0 0 0 0
39 41 1 0 0 0 0
35 39 1 0 0 0 0
42 43 2 0 0 0 0
42 44 1 0 0 0 0
24 42 1 0 0 0 0
M STY 1 1 SUP
M SLB 1 1 1
M SAL 1 3 39 40 41
M SBL 1 1 45
M SMT 1 ^COOH
M SBV 1 45 0.5604 -0.3973
M STY 1 2 SUP
M SLB 1 2 2
M SAL 2 3 42 43 44
M SBL 2 1 48
M SMT 2 COOH
M SBV 2 48 -0.7758 0.4479
S SKP 5
ID FL3FAAGS0027
FORMULA C27H26O17
EXACTMASS 622.116999406
AVERAGEMASS 622.48514
SMILES C(Oc(c2)ccc(C(=C5)Oc(c3)c(C5=O)c(cc3OC(O4)C(C(C(O)C4C(O)=O)O)O)O)c2)(O1)C(C(C(O)C1C(O)=O)O)O
M END