Mol:FL3FAACS0062
From Metabolomics.JP
(Difference between revisions)
| Line 1: | Line 1: | ||
| − | + | ||
| − | + | ||
| − | Copyright: ARM project http://www.metabolome.jp/ | + | Copyright: ARM project http://www.metabolome.jp/ |
| − | 53 58 0 0 0 0 0 0 0 0999 V2000 | + | 53 58 0 0 0 0 0 0 0 0999 V2000 |
| − | 0.2734 -0.9637 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.2734 -0.9637 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.2734 -1.6061 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.2734 -1.6061 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.8297 -1.9273 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.8297 -1.9273 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.3860 -1.6061 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.3860 -1.6061 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.3860 -0.9637 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.3860 -0.9637 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.8297 -0.6425 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.8297 -0.6425 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.9423 -1.9273 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.9423 -1.9273 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.4986 -1.6061 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.4986 -1.6061 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.4986 -0.9637 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.4986 -0.9637 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.9423 -0.6425 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.9423 -0.6425 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.9423 -2.4281 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.9423 -2.4281 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.2827 -0.6427 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.2827 -0.6427 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.0751 2.0419 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.0751 2.0419 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.4695 1.5831 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.4695 1.5831 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.7264 0.9225 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.7264 0.9225 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.7333 0.2850 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.7333 0.2850 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.1966 0.7483 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.1966 0.7483 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.8915 1.3262 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.8915 1.3262 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.6765 2.7323 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.6765 2.7323 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.4347 2.2983 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.4347 2.2983 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.0909 0.5440 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.0909 0.5440 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.8297 -2.5694 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.8297 -2.5694 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.1165 -0.6220 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.1165 -0.6220 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.7027 -0.9605 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.7027 -0.9605 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.2890 -0.6220 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.2890 -0.6220 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.2890 0.0549 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.2890 0.0549 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.7027 0.3933 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.7027 0.3933 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.1165 0.0549 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.1165 0.0549 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.8748 0.3931 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.8748 0.3931 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.8241 -1.9361 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.8241 -1.9361 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.4529 -2.4260 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.4529 -2.4260 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.9184 -2.2182 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.9184 -2.2182 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.4027 -2.2126 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.4027 -2.2126 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.7775 -1.8378 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.7775 -1.8378 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.2451 -2.0846 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.2451 -2.0846 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.3379 -2.2328 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.3379 -2.2328 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.0316 -2.4542 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.0316 -2.4542 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.6122 -2.7323 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.6122 -2.7323 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.7685 -1.3762 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.7685 -1.3762 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.3610 -1.3792 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.3610 -1.3792 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.9898 -1.8692 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.9898 -1.8692 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.4553 -1.6613 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.4553 -1.6613 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.9396 -1.6557 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.9396 -1.6557 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.3143 -1.2809 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.3143 -1.2809 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.7819 -1.5277 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.7819 -1.5277 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.8748 -1.6759 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.8748 -1.6759 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.5684 -1.8973 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.5684 -1.8973 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.1491 -2.1754 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.1491 -2.1754 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.4491 -1.2937 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.4491 -1.2937 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.3152 1.5708 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.3152 1.5708 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.0297 1.1583 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.0297 1.1583 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.1804 -0.7435 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.1804 -0.7435 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.3835 -0.9570 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.3835 -0.9570 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1 2 1 0 0 0 0 | + | 1 2 1 0 0 0 0 |
| − | 2 3 2 0 0 0 0 | + | 2 3 2 0 0 0 0 |
| − | 3 4 1 0 0 0 0 | + | 3 4 1 0 0 0 0 |
| − | 4 5 2 0 0 0 0 | + | 4 5 2 0 0 0 0 |
| − | 5 6 1 0 0 0 0 | + | 5 6 1 0 0 0 0 |
| − | 6 1 2 0 0 0 0 | + | 6 1 2 0 0 0 0 |
| − | 4 7 1 0 0 0 0 | + | 4 7 1 0 0 0 0 |
| − | 7 8 1 0 0 0 0 | + | 7 8 1 0 0 0 0 |
| − | 8 9 2 0 0 0 0 | + | 8 9 2 0 0 0 0 |
| − | 9 10 1 0 0 0 0 | + | 9 10 1 0 0 0 0 |
| − | 10 5 1 0 0 0 0 | + | 10 5 1 0 0 0 0 |
| − | 7 11 2 0 0 0 0 | + | 7 11 2 0 0 0 0 |
| − | 1 12 1 0 0 0 0 | + | 1 12 1 0 0 0 0 |
| − | 13 14 1 1 0 0 0 | + | 13 14 1 1 0 0 0 |
| − | 14 15 1 1 0 0 0 | + | 14 15 1 1 0 0 0 |
| − | 16 15 1 1 0 0 0 | + | 16 15 1 1 0 0 0 |
| − | 16 17 1 0 0 0 0 | + | 16 17 1 0 0 0 0 |
| − | 17 18 1 0 0 0 0 | + | 17 18 1 0 0 0 0 |
| − | 18 13 1 0 0 0 0 | + | 18 13 1 0 0 0 0 |
| − | 13 19 1 0 0 0 0 | + | 13 19 1 0 0 0 0 |
| − | 14 20 1 0 0 0 0 | + | 14 20 1 0 0 0 0 |
| − | 15 21 1 0 0 0 0 | + | 15 21 1 0 0 0 0 |
| − | 6 16 1 0 0 0 0 | + | 6 16 1 0 0 0 0 |
| − | 3 22 1 0 0 0 0 | + | 3 22 1 0 0 0 0 |
| − | 23 24 2 0 0 0 0 | + | 23 24 2 0 0 0 0 |
| − | 24 25 1 0 0 0 0 | + | 24 25 1 0 0 0 0 |
| − | 25 26 2 0 0 0 0 | + | 25 26 2 0 0 0 0 |
| − | 26 27 1 0 0 0 0 | + | 26 27 1 0 0 0 0 |
| − | 27 28 2 0 0 0 0 | + | 27 28 2 0 0 0 0 |
| − | 28 23 1 0 0 0 0 | + | 28 23 1 0 0 0 0 |
| − | 23 9 1 0 0 0 0 | + | 23 9 1 0 0 0 0 |
| − | 26 29 1 0 0 0 0 | + | 26 29 1 0 0 0 0 |
| − | 30 31 1 1 0 0 0 | + | 30 31 1 1 0 0 0 |
| − | 31 32 1 1 0 0 0 | + | 31 32 1 1 0 0 0 |
| − | 33 32 1 1 0 0 0 | + | 33 32 1 1 0 0 0 |
| − | 33 34 1 0 0 0 0 | + | 33 34 1 0 0 0 0 |
| − | 34 35 1 0 0 0 0 | + | 34 35 1 0 0 0 0 |
| − | 35 30 1 0 0 0 0 | + | 35 30 1 0 0 0 0 |
| − | 30 36 1 0 0 0 0 | + | 30 36 1 0 0 0 0 |
| − | 31 37 1 0 0 0 0 | + | 31 37 1 0 0 0 0 |
| − | 32 38 1 0 0 0 0 | + | 32 38 1 0 0 0 0 |
| − | 35 39 1 0 0 0 0 | + | 35 39 1 0 0 0 0 |
| − | 33 2 1 0 0 0 0 | + | 33 2 1 0 0 0 0 |
| − | 40 41 1 1 0 0 0 | + | 40 41 1 1 0 0 0 |
| − | 41 42 1 1 0 0 0 | + | 41 42 1 1 0 0 0 |
| − | 43 42 1 1 0 0 0 | + | 43 42 1 1 0 0 0 |
| − | 43 44 1 0 0 0 0 | + | 43 44 1 0 0 0 0 |
| − | 44 45 1 0 0 0 0 | + | 44 45 1 0 0 0 0 |
| − | 45 40 1 0 0 0 0 | + | 45 40 1 0 0 0 0 |
| − | 40 46 1 0 0 0 0 | + | 40 46 1 0 0 0 0 |
| − | 41 47 1 0 0 0 0 | + | 41 47 1 0 0 0 0 |
| − | 42 48 1 0 0 0 0 | + | 42 48 1 0 0 0 0 |
| − | 43 49 1 0 0 0 0 | + | 43 49 1 0 0 0 0 |
| − | 49 39 1 0 0 0 0 | + | 49 39 1 0 0 0 0 |
| − | 18 50 1 0 0 0 0 | + | 18 50 1 0 0 0 0 |
| − | 50 51 1 0 0 0 0 | + | 50 51 1 0 0 0 0 |
| − | 45 52 1 0 0 0 0 | + | 45 52 1 0 0 0 0 |
| − | 52 53 1 0 0 0 0 | + | 52 53 1 0 0 0 0 |
| − | M STY 1 1 SUP | + | M STY 1 1 SUP |
| − | M SLB 1 1 1 | + | M SLB 1 1 1 |
| − | M SAL 1 2 50 51 | + | M SAL 1 2 50 51 |
| − | M SBL 1 1 55 | + | M SBL 1 1 55 |
| − | M SMT 1 CH2OH | + | M SMT 1 CH2OH |
| − | M SBV 1 55 -4.9501 5.5921 | + | M SBV 1 55 -4.9501 5.5921 |
| − | M STY 1 2 SUP | + | M STY 1 2 SUP |
| − | M SLB 1 2 2 | + | M SLB 1 2 2 |
| − | M SAL 2 2 52 53 | + | M SAL 2 2 52 53 |
| − | M SBL 2 1 57 | + | M SBL 2 1 57 |
| − | M SMT 2 ^CH2OH | + | M SMT 2 ^CH2OH |
| − | M SBV 2 57 -5.7722 6.1317 | + | M SBV 2 57 -5.7722 6.1317 |
| − | S SKP 8 | + | S SKP 8 |
| − | ID FL3FAACS0062 | + | ID FL3FAACS0062 |
| − | KNApSAcK_ID C00006408 | + | KNApSAcK_ID C00006408 |
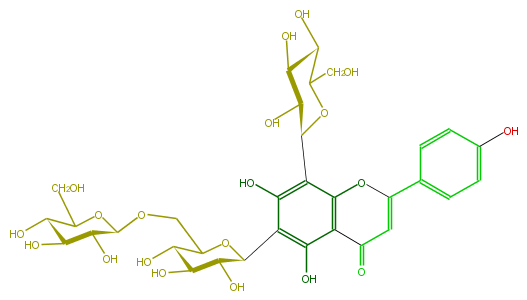
| − | NAME Vicenin-2,6''-O-glucoside | + | NAME Vicenin-2,6''-O-glucoside |
| − | CAS_RN 94530-40-2 | + | CAS_RN 94530-40-2 |
| − | FORMULA C33H40O20 | + | FORMULA C33H40O20 |
| − | EXACTMASS 756.21129372 | + | EXACTMASS 756.21129372 |
| − | AVERAGEMASS 756.6587 | + | AVERAGEMASS 756.6587 |
| − | SMILES OC(C(O)1)C(CO)OC(c(c42)c(c(C(O5)C(C(O)C(C5COC(O6)C(O)C(C(C(CO)6)O)O)O)O)c(O)c2C(=O)C=C(O4)c(c3)ccc(O)c3)O)C(O)1 | + | SMILES OC(C(O)1)C(CO)OC(c(c42)c(c(C(O5)C(C(O)C(C5COC(O6)C(O)C(C(C(CO)6)O)O)O)O)c(O)c2C(=O)C=C(O4)c(c3)ccc(O)c3)O)C(O)1 |
M END | M END | ||
| − | |||
Latest revision as of 09:00, 14 March 2009
Copyright: ARM project http://www.metabolome.jp/
53 58 0 0 0 0 0 0 0 0999 V2000
0.2734 -0.9637 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.2734 -1.6061 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.8297 -1.9273 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.3860 -1.6061 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.3860 -0.9637 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.8297 -0.6425 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.9423 -1.9273 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.4986 -1.6061 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.4986 -0.9637 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.9423 -0.6425 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.9423 -2.4281 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.2827 -0.6427 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.0751 2.0419 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.4695 1.5831 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.7264 0.9225 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.7333 0.2850 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.1966 0.7483 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.8915 1.3262 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.6765 2.7323 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.4347 2.2983 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.0909 0.5440 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.8297 -2.5694 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.1165 -0.6220 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.7027 -0.9605 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.2890 -0.6220 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.2890 0.0549 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.7027 0.3933 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.1165 0.0549 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.8748 0.3931 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.8241 -1.9361 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.4529 -2.4260 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.9184 -2.2182 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.4027 -2.2126 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.7775 -1.8378 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.2451 -2.0846 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.3379 -2.2328 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.0316 -2.4542 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.6122 -2.7323 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.7685 -1.3762 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.3610 -1.3792 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.9898 -1.8692 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.4553 -1.6613 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.9396 -1.6557 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.3143 -1.2809 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.7819 -1.5277 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.8748 -1.6759 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.5684 -1.8973 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.1491 -2.1754 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.4491 -1.2937 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.3152 1.5708 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.0297 1.1583 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.1804 -0.7435 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.3835 -0.9570 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1 2 1 0 0 0 0
2 3 2 0 0 0 0
3 4 1 0 0 0 0
4 5 2 0 0 0 0
5 6 1 0 0 0 0
6 1 2 0 0 0 0
4 7 1 0 0 0 0
7 8 1 0 0 0 0
8 9 2 0 0 0 0
9 10 1 0 0 0 0
10 5 1 0 0 0 0
7 11 2 0 0 0 0
1 12 1 0 0 0 0
13 14 1 1 0 0 0
14 15 1 1 0 0 0
16 15 1 1 0 0 0
16 17 1 0 0 0 0
17 18 1 0 0 0 0
18 13 1 0 0 0 0
13 19 1 0 0 0 0
14 20 1 0 0 0 0
15 21 1 0 0 0 0
6 16 1 0 0 0 0
3 22 1 0 0 0 0
23 24 2 0 0 0 0
24 25 1 0 0 0 0
25 26 2 0 0 0 0
26 27 1 0 0 0 0
27 28 2 0 0 0 0
28 23 1 0 0 0 0
23 9 1 0 0 0 0
26 29 1 0 0 0 0
30 31 1 1 0 0 0
31 32 1 1 0 0 0
33 32 1 1 0 0 0
33 34 1 0 0 0 0
34 35 1 0 0 0 0
35 30 1 0 0 0 0
30 36 1 0 0 0 0
31 37 1 0 0 0 0
32 38 1 0 0 0 0
35 39 1 0 0 0 0
33 2 1 0 0 0 0
40 41 1 1 0 0 0
41 42 1 1 0 0 0
43 42 1 1 0 0 0
43 44 1 0 0 0 0
44 45 1 0 0 0 0
45 40 1 0 0 0 0
40 46 1 0 0 0 0
41 47 1 0 0 0 0
42 48 1 0 0 0 0
43 49 1 0 0 0 0
49 39 1 0 0 0 0
18 50 1 0 0 0 0
50 51 1 0 0 0 0
45 52 1 0 0 0 0
52 53 1 0 0 0 0
M STY 1 1 SUP
M SLB 1 1 1
M SAL 1 2 50 51
M SBL 1 1 55
M SMT 1 CH2OH
M SBV 1 55 -4.9501 5.5921
M STY 1 2 SUP
M SLB 1 2 2
M SAL 2 2 52 53
M SBL 2 1 57
M SMT 2 ^CH2OH
M SBV 2 57 -5.7722 6.1317
S SKP 8
ID FL3FAACS0062
KNApSAcK_ID C00006408
NAME Vicenin-2,6''-O-glucoside
CAS_RN 94530-40-2
FORMULA C33H40O20
EXACTMASS 756.21129372
AVERAGEMASS 756.6587
SMILES OC(C(O)1)C(CO)OC(c(c42)c(c(C(O5)C(C(O)C(C5COC(O6)C(O)C(C(C(CO)6)O)O)O)O)c(O)c2C(=O)C=C(O4)c(c3)ccc(O)c3)O)C(O)1
M END