Mol:FLID1CGS0005
From Metabolomics.JP
(Difference between revisions)
| Line 1: | Line 1: | ||
| − | + | ||
| − | + | ||
| − | Copyright: ARM project http://www.metabolome.jp/ | + | Copyright: ARM project http://www.metabolome.jp/ |
| − | 33 37 0 0 0 0 0 0 0 0999 V2000 | + | 33 37 0 0 0 0 0 0 0 0999 V2000 |
| − | -1.2121 1.1949 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.2121 1.1949 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.6558 0.8737 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.6558 0.8737 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.0995 1.1949 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.0995 1.1949 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.4566 0.8738 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.4566 0.8738 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.4566 0.2079 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.4566 0.2079 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.0333 -0.1251 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 | + | 1.0333 -0.1251 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 |
| − | 1.6101 0.2079 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 | + | 1.6101 0.2079 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 |
| − | 1.6101 0.8738 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.6101 0.8738 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.0333 1.2068 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.0333 1.2068 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.6556 0.2317 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.6556 0.2317 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.0995 -0.0894 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.0995 -0.0894 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.0372 -0.7615 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.0372 -0.7615 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.1864 -0.1249 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.1864 -0.1249 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.1864 -0.7615 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.1864 -0.7615 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.7378 -1.0799 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.7378 -1.0799 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.2891 -0.7615 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.2891 -0.7615 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.2891 -0.1249 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.2891 -0.1249 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.7378 0.1934 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.7378 0.1934 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.3573 0.8261 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0 | + | -3.3573 0.8261 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0 |
| − | -3.0110 0.3691 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | -3.0110 0.3691 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | -2.5124 0.5630 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | -2.5124 0.5630 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | -1.9928 0.5572 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | -1.9928 0.5572 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | -2.3809 0.9178 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.3809 0.9178 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.8902 0.7350 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 | + | -2.8902 0.7350 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 |
| − | -4.1977 0.6009 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.1977 0.6009 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.5508 0.3428 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.5508 0.3428 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.2268 0.0834 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.2268 0.0834 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.2891 -0.7615 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.2891 -0.7615 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 5.2891 -0.7615 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 5.2891 -0.7615 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.2989 -1.2390 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.2989 -1.2390 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.0134 -1.6515 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.0134 -1.6515 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.4639 1.4787 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.4639 1.4787 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.8763 2.1932 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.8763 2.1932 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1 2 1 0 0 0 0 | + | 1 2 1 0 0 0 0 |
| − | 2 3 2 0 0 0 0 | + | 2 3 2 0 0 0 0 |
| − | 3 4 1 0 0 0 0 | + | 3 4 1 0 0 0 0 |
| − | 4 5 2 0 0 0 0 | + | 4 5 2 0 0 0 0 |
| − | 5 6 1 0 0 0 0 | + | 5 6 1 0 0 0 0 |
| − | 6 7 1 0 0 0 0 | + | 6 7 1 0 0 0 0 |
| − | 7 8 1 0 0 0 0 | + | 7 8 1 0 0 0 0 |
| − | 8 9 1 0 0 0 0 | + | 8 9 1 0 0 0 0 |
| − | 9 4 1 0 0 0 0 | + | 9 4 1 0 0 0 0 |
| − | 2 10 1 0 0 0 0 | + | 2 10 1 0 0 0 0 |
| − | 10 11 2 0 0 0 0 | + | 10 11 2 0 0 0 0 |
| − | 11 5 1 0 0 0 0 | + | 11 5 1 0 0 0 0 |
| − | 6 12 1 0 0 0 0 | + | 6 12 1 0 0 0 0 |
| − | 7 13 1 0 0 0 0 | + | 7 13 1 0 0 0 0 |
| − | 13 14 2 0 0 0 0 | + | 13 14 2 0 0 0 0 |
| − | 14 15 1 0 0 0 0 | + | 14 15 1 0 0 0 0 |
| − | 15 16 2 0 0 0 0 | + | 15 16 2 0 0 0 0 |
| − | 16 17 1 0 0 0 0 | + | 16 17 1 0 0 0 0 |
| − | 17 18 2 0 0 0 0 | + | 17 18 2 0 0 0 0 |
| − | 18 13 1 0 0 0 0 | + | 18 13 1 0 0 0 0 |
| − | 19 20 1 1 0 0 0 | + | 19 20 1 1 0 0 0 |
| − | 20 21 1 1 0 0 0 | + | 20 21 1 1 0 0 0 |
| − | 22 21 1 1 0 0 0 | + | 22 21 1 1 0 0 0 |
| − | 22 23 1 0 0 0 0 | + | 22 23 1 0 0 0 0 |
| − | 23 24 1 0 0 0 0 | + | 23 24 1 0 0 0 0 |
| − | 24 19 1 0 0 0 0 | + | 24 19 1 0 0 0 0 |
| − | 19 25 1 0 0 0 0 | + | 19 25 1 0 0 0 0 |
| − | 20 26 1 0 0 0 0 | + | 20 26 1 0 0 0 0 |
| − | 21 27 1 0 0 0 0 | + | 21 27 1 0 0 0 0 |
| − | 22 1 1 0 0 0 0 | + | 22 1 1 0 0 0 0 |
| − | 14 12 1 0 0 0 0 | + | 14 12 1 0 0 0 0 |
| − | 16 28 1 0 0 0 0 | + | 16 28 1 0 0 0 0 |
| − | 28 29 1 0 0 0 0 | + | 28 29 1 0 0 0 0 |
| − | 15 30 1 0 0 0 0 | + | 15 30 1 0 0 0 0 |
| − | 30 31 1 0 0 0 0 | + | 30 31 1 0 0 0 0 |
| − | 24 32 1 0 0 0 0 | + | 24 32 1 0 0 0 0 |
| − | 32 33 1 0 0 0 0 | + | 32 33 1 0 0 0 0 |
| − | M STY 1 3 SUP | + | M STY 1 3 SUP |
| − | M SLB 1 3 3 | + | M SLB 1 3 3 |
| − | M SAL 3 2 32 33 | + | M SAL 3 2 32 33 |
| − | M SBL 3 1 36 | + | M SBL 3 1 36 |
| − | M SMT 3 CH2OH | + | M SMT 3 CH2OH |
| − | M SVB 3 36 -3.0958 1.6515 | + | M SVB 3 36 -3.0958 1.6515 |
| − | M STY 1 2 SUP | + | M STY 1 2 SUP |
| − | M SLB 1 2 2 | + | M SLB 1 2 2 |
| − | M SAL 2 2 30 31 | + | M SAL 2 2 30 31 |
| − | M SBL 2 1 34 | + | M SBL 2 1 34 |
| − | M SMT 2 OCH3 | + | M SMT 2 OCH3 |
| − | M SVB 2 34 2.2989 -1.239 | + | M SVB 2 34 2.2989 -1.239 |
| − | M STY 1 1 SUP | + | M STY 1 1 SUP |
| − | M SLB 1 1 1 | + | M SLB 1 1 1 |
| − | M SAL 1 2 28 29 | + | M SAL 1 2 28 29 |
| − | M SBL 1 1 32 | + | M SBL 1 1 32 |
| − | M SMT 1 OCH3 | + | M SMT 1 OCH3 |
| − | M SVB 1 32 3.4832 -0.9678 | + | M SVB 1 32 3.4832 -0.9678 |
| − | S SKP 8 | + | S SKP 8 |
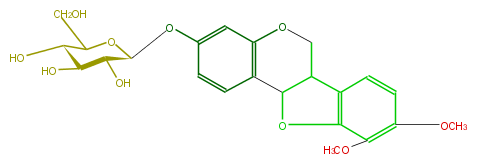
| − | ID FLID1CGS0005 | + | ID FLID1CGS0005 |
| − | KNApSAcK_ID C00010189 | + | KNApSAcK_ID C00010189 |
| − | NAME Methylnissolin 3-O-glucoside | + | NAME Methylnissolin 3-O-glucoside |
| − | CAS_RN 94367-42-7 | + | CAS_RN 94367-42-7 |
| − | FORMULA C23H26O10 | + | FORMULA C23H26O10 |
| − | EXACTMASS 462.152597052 | + | EXACTMASS 462.152597052 |
| − | AVERAGEMASS 462.44654 | + | AVERAGEMASS 462.44654 |
| − | SMILES COc(c51)c(ccc(C(C4)C(O5)c(c3)c(O4)cc(c3)O[C@@H]([C@@H](O)2)OC(CO)[C@H](O)[C@@H]2O)1)OC | + | SMILES COc(c51)c(ccc(C(C4)C(O5)c(c3)c(O4)cc(c3)O[C@@H]([C@@H](O)2)OC(CO)[C@H](O)[C@@H]2O)1)OC |
M END | M END | ||
| − | |||
Latest revision as of 09:00, 14 March 2009
Copyright: ARM project http://www.metabolome.jp/
33 37 0 0 0 0 0 0 0 0999 V2000
-1.2121 1.1949 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.6558 0.8737 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.0995 1.1949 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.4566 0.8738 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.4566 0.2079 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.0333 -0.1251 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0
1.6101 0.2079 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0
1.6101 0.8738 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.0333 1.2068 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.6556 0.2317 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.0995 -0.0894 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.0372 -0.7615 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.1864 -0.1249 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.1864 -0.7615 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.7378 -1.0799 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.2891 -0.7615 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.2891 -0.1249 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.7378 0.1934 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.3573 0.8261 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0
-3.0110 0.3691 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
-2.5124 0.5630 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
-1.9928 0.5572 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
-2.3809 0.9178 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.8902 0.7350 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0
-4.1977 0.6009 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.5508 0.3428 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.2268 0.0834 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
4.2891 -0.7615 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
5.2891 -0.7615 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.2989 -1.2390 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.0134 -1.6515 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.4639 1.4787 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.8763 2.1932 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1 2 1 0 0 0 0
2 3 2 0 0 0 0
3 4 1 0 0 0 0
4 5 2 0 0 0 0
5 6 1 0 0 0 0
6 7 1 0 0 0 0
7 8 1 0 0 0 0
8 9 1 0 0 0 0
9 4 1 0 0 0 0
2 10 1 0 0 0 0
10 11 2 0 0 0 0
11 5 1 0 0 0 0
6 12 1 0 0 0 0
7 13 1 0 0 0 0
13 14 2 0 0 0 0
14 15 1 0 0 0 0
15 16 2 0 0 0 0
16 17 1 0 0 0 0
17 18 2 0 0 0 0
18 13 1 0 0 0 0
19 20 1 1 0 0 0
20 21 1 1 0 0 0
22 21 1 1 0 0 0
22 23 1 0 0 0 0
23 24 1 0 0 0 0
24 19 1 0 0 0 0
19 25 1 0 0 0 0
20 26 1 0 0 0 0
21 27 1 0 0 0 0
22 1 1 0 0 0 0
14 12 1 0 0 0 0
16 28 1 0 0 0 0
28 29 1 0 0 0 0
15 30 1 0 0 0 0
30 31 1 0 0 0 0
24 32 1 0 0 0 0
32 33 1 0 0 0 0
M STY 1 3 SUP
M SLB 1 3 3
M SAL 3 2 32 33
M SBL 3 1 36
M SMT 3 CH2OH
M SVB 3 36 -3.0958 1.6515
M STY 1 2 SUP
M SLB 1 2 2
M SAL 2 2 30 31
M SBL 2 1 34
M SMT 2 OCH3
M SVB 2 34 2.2989 -1.239
M STY 1 1 SUP
M SLB 1 1 1
M SAL 1 2 28 29
M SBL 1 1 32
M SMT 1 OCH3
M SVB 1 32 3.4832 -0.9678
S SKP 8
ID FLID1CGS0005
KNApSAcK_ID C00010189
NAME Methylnissolin 3-O-glucoside
CAS_RN 94367-42-7
FORMULA C23H26O10
EXACTMASS 462.152597052
AVERAGEMASS 462.44654
SMILES COc(c51)c(ccc(C(C4)C(O5)c(c3)c(O4)cc(c3)O[C@@H]([C@@H](O)2)OC(CO)[C@H](O)[C@@H]2O)1)OC
M END