Mol:FLIAEAGS0008
From Metabolomics.JP
(Difference between revisions)
| Line 1: | Line 1: | ||
| − | + | ||
| − | + | ||
| − | Copyright: ARM project http://www.metabolome.jp/ | + | Copyright: ARM project http://www.metabolome.jp/ |
| − | 46 50 0 0 0 0 0 0 0 0999 V2000 | + | 46 50 0 0 0 0 0 0 0 0999 V2000 |
| − | -0.2530 1.0593 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.2530 1.0593 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.3033 0.7381 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.3033 0.7381 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.8596 1.0593 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.8596 1.0593 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.4157 0.7382 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.4157 0.7382 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.4157 0.0722 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.4157 0.0722 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.9924 -0.2607 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.9924 -0.2607 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.5691 0.0722 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.5691 0.0722 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.5691 0.7382 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.5691 0.7382 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.9924 1.0712 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.9924 1.0712 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.3035 0.0961 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.3035 0.0961 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.8596 -0.2250 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.8596 -0.2250 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.1455 -0.2605 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.1455 -0.2605 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.1455 -0.8971 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.1455 -0.8971 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.6968 -1.2155 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.6968 -1.2155 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.2482 -0.8971 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.2482 -0.8971 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.2482 -0.2605 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.2482 -0.2605 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.6968 0.0578 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.6968 0.0578 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.3982 0.6905 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0 | + | -2.3982 0.6905 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0 |
| − | -2.0520 0.2335 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | -2.0520 0.2335 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | -1.5534 0.4274 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | -1.5534 0.4274 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | -1.0337 0.4216 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | -1.0337 0.4216 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | -1.4219 0.7822 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.4219 0.7822 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.9312 0.5993 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 | + | -1.9312 0.5993 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 |
| − | -3.2386 0.4653 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.2386 0.4653 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.5917 0.2072 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.5917 0.2072 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.2677 -0.0522 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.2677 -0.0522 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.3163 -0.4851 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0 | + | -4.3163 -0.4851 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0 |
| − | -3.9701 -0.9422 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | -3.9701 -0.9422 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | -3.4715 -0.7483 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | -3.4715 -0.7483 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | -2.9518 -0.7540 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | -2.9518 -0.7540 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | -3.3400 -0.3934 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.3400 -0.3934 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.8493 -0.5763 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 | + | -3.8493 -0.5763 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 |
| − | -5.1567 -0.7103 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -5.1567 -0.7103 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.5098 -0.9684 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.5098 -0.9684 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.1858 -1.2279 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.1858 -1.2279 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.9924 -0.9262 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.9924 -0.9262 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 5.1143 -1.3970 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 5.1143 -1.3970 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 5.9803 -1.8970 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 5.9803 -1.8970 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.5049 1.3431 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.5049 1.3431 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.5768 2.1650 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.5768 2.1650 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.8597 -1.2250 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.8597 -1.2250 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.8597 -2.2250 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.8597 -2.2250 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.3370 -0.6235 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.3370 -0.6235 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.3701 -1.3306 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.3701 -1.3306 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.6836 -0.1446 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.6836 -0.1446 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -5.0961 0.5699 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -5.0961 0.5699 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1 2 1 0 0 0 0 | + | 1 2 1 0 0 0 0 |
| − | 2 3 2 0 0 0 0 | + | 2 3 2 0 0 0 0 |
| − | 3 4 1 0 0 0 0 | + | 3 4 1 0 0 0 0 |
| − | 4 5 2 0 0 0 0 | + | 4 5 2 0 0 0 0 |
| − | 5 6 1 0 0 0 0 | + | 5 6 1 0 0 0 0 |
| − | 6 7 1 0 0 0 0 | + | 6 7 1 0 0 0 0 |
| − | 7 8 2 0 0 0 0 | + | 7 8 2 0 0 0 0 |
| − | 8 9 1 0 0 0 0 | + | 8 9 1 0 0 0 0 |
| − | 9 4 1 0 0 0 0 | + | 9 4 1 0 0 0 0 |
| − | 2 10 1 0 0 0 0 | + | 2 10 1 0 0 0 0 |
| − | 10 11 2 0 0 0 0 | + | 10 11 2 0 0 0 0 |
| − | 11 5 1 0 0 0 0 | + | 11 5 1 0 0 0 0 |
| − | 7 12 1 0 0 0 0 | + | 7 12 1 0 0 0 0 |
| − | 12 13 2 0 0 0 0 | + | 12 13 2 0 0 0 0 |
| − | 13 14 1 0 0 0 0 | + | 13 14 1 0 0 0 0 |
| − | 14 15 2 0 0 0 0 | + | 14 15 2 0 0 0 0 |
| − | 15 16 1 0 0 0 0 | + | 15 16 1 0 0 0 0 |
| − | 16 17 2 0 0 0 0 | + | 16 17 2 0 0 0 0 |
| − | 17 12 1 0 0 0 0 | + | 17 12 1 0 0 0 0 |
| − | 18 19 1 1 0 0 0 | + | 18 19 1 1 0 0 0 |
| − | 19 20 1 1 0 0 0 | + | 19 20 1 1 0 0 0 |
| − | 21 20 1 1 0 0 0 | + | 21 20 1 1 0 0 0 |
| − | 21 22 1 0 0 0 0 | + | 21 22 1 0 0 0 0 |
| − | 22 23 1 0 0 0 0 | + | 22 23 1 0 0 0 0 |
| − | 23 18 1 0 0 0 0 | + | 23 18 1 0 0 0 0 |
| − | 18 24 1 0 0 0 0 | + | 18 24 1 0 0 0 0 |
| − | 19 25 1 0 0 0 0 | + | 19 25 1 0 0 0 0 |
| − | 20 26 1 0 0 0 0 | + | 20 26 1 0 0 0 0 |
| − | 21 1 1 0 0 0 0 | + | 21 1 1 0 0 0 0 |
| − | 27 28 1 1 0 0 0 | + | 27 28 1 1 0 0 0 |
| − | 28 29 1 1 0 0 0 | + | 28 29 1 1 0 0 0 |
| − | 30 29 1 1 0 0 0 | + | 30 29 1 1 0 0 0 |
| − | 30 31 1 0 0 0 0 | + | 30 31 1 0 0 0 0 |
| − | 31 32 1 0 0 0 0 | + | 31 32 1 0 0 0 0 |
| − | 32 27 1 0 0 0 0 | + | 32 27 1 0 0 0 0 |
| − | 27 33 1 0 0 0 0 | + | 27 33 1 0 0 0 0 |
| − | 28 34 1 0 0 0 0 | + | 28 34 1 0 0 0 0 |
| − | 29 35 1 0 0 0 0 | + | 29 35 1 0 0 0 0 |
| − | 30 25 1 0 0 0 0 | + | 30 25 1 0 0 0 0 |
| − | 6 36 2 0 0 0 0 | + | 6 36 2 0 0 0 0 |
| − | 15 37 1 0 0 0 0 | + | 15 37 1 0 0 0 0 |
| − | 37 38 1 0 0 0 0 | + | 37 38 1 0 0 0 0 |
| − | 23 39 1 0 0 0 0 | + | 23 39 1 0 0 0 0 |
| − | 39 40 1 0 0 0 0 | + | 39 40 1 0 0 0 0 |
| − | 11 41 1 0 0 0 0 | + | 11 41 1 0 0 0 0 |
| − | 41 42 1 0 0 0 0 | + | 41 42 1 0 0 0 0 |
| − | 10 43 1 0 0 0 0 | + | 10 43 1 0 0 0 0 |
| − | 43 44 1 0 0 0 0 | + | 43 44 1 0 0 0 0 |
| − | 32 45 1 0 0 0 0 | + | 32 45 1 0 0 0 0 |
| − | 45 46 1 0 0 0 0 | + | 45 46 1 0 0 0 0 |
| − | M STY 1 5 SUP | + | M STY 1 5 SUP |
| − | M SLB 1 5 5 | + | M SLB 1 5 5 |
| − | M SAL 5 2 45 46 | + | M SAL 5 2 45 46 |
| − | M SBL 5 1 49 | + | M SBL 5 1 49 |
| − | M SMT 5 CH2OH | + | M SMT 5 CH2OH |
| − | M SVB 5 49 -4.0549 0.3403 | + | M SVB 5 49 -4.0549 0.3403 |
| − | M STY 1 4 SUP | + | M STY 1 4 SUP |
| − | M SLB 1 4 4 | + | M SLB 1 4 4 |
| − | M SAL 4 2 39 40 | + | M SAL 4 2 39 40 |
| − | M SBL 4 1 43 | + | M SBL 4 1 43 |
| − | M SMT 4 CH2OH | + | M SMT 4 CH2OH |
| − | M SVB 4 43 -2.1368 1.5159 | + | M SVB 4 43 -2.1368 1.5159 |
| − | M STY 1 3 SUP | + | M STY 1 3 SUP |
| − | M SLB 1 3 3 | + | M SLB 1 3 3 |
| − | M SAL 3 2 43 44 | + | M SAL 3 2 43 44 |
| − | M SBL 3 1 47 | + | M SBL 3 1 47 |
| − | M SMT 3 OCH3 | + | M SMT 3 OCH3 |
| − | M SVB 3 47 -0.411 0.1881 | + | M SVB 3 47 -0.411 0.1881 |
| − | M STY 1 2 SUP | + | M STY 1 2 SUP |
| − | M SLB 1 2 2 | + | M SLB 1 2 2 |
| − | M SAL 2 2 41 42 | + | M SAL 2 2 41 42 |
| − | M SBL 2 1 45 | + | M SBL 2 1 45 |
| − | M SMT 2 OCH3 | + | M SMT 2 OCH3 |
| − | M SVB 2 45 0.4363 -0.3931 | + | M SVB 2 45 0.4363 -0.3931 |
| − | M STY 1 1 SUP | + | M STY 1 1 SUP |
| − | M SLB 1 1 1 | + | M SLB 1 1 1 |
| − | M SAL 1 2 37 38 | + | M SAL 1 2 37 38 |
| − | M SBL 1 1 41 | + | M SBL 1 1 41 |
| − | M SMT 1 OCH3 | + | M SMT 1 OCH3 |
| − | M SVB 1 41 4.4423 -1.1034 | + | M SVB 1 41 4.4423 -1.1034 |
| − | S SKP 8 | + | S SKP 8 |
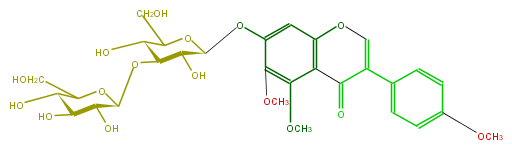
| − | ID FLIAEAGS0008 | + | ID FLIAEAGS0008 |
| − | KNApSAcK_ID C00010130 | + | KNApSAcK_ID C00010130 |
| − | NAME 5-Methoxyafrormosin 7-O-laminaribioside | + | NAME 5-Methoxyafrormosin 7-O-laminaribioside |
| − | CAS_RN 68862-18-0 | + | CAS_RN 68862-18-0 |
| − | FORMULA C30H36O16 | + | FORMULA C30H36O16 |
| − | EXACTMASS 652.200335104 | + | EXACTMASS 652.200335104 |
| − | AVERAGEMASS 652.59724 | + | AVERAGEMASS 652.59724 |
| − | SMILES O[C@@H]([C@H]4O[C@@H]([C@@H](O)5)OC([C@H](O)[C@@H]5O)CO)[C@@H](OC(CO)[C@@H]4O)Oc(c3)c(c(OC)c(c23)C(C(=CO2)c(c1)ccc(OC)c1)=O)OC | + | SMILES O[C@@H]([C@H]4O[C@@H]([C@@H](O)5)OC([C@H](O)[C@@H]5O)CO)[C@@H](OC(CO)[C@@H]4O)Oc(c3)c(c(OC)c(c23)C(C(=CO2)c(c1)ccc(OC)c1)=O)OC |
M END | M END | ||
| − | |||
Latest revision as of 09:00, 14 March 2009
Copyright: ARM project http://www.metabolome.jp/
46 50 0 0 0 0 0 0 0 0999 V2000
-0.2530 1.0593 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.3033 0.7381 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.8596 1.0593 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.4157 0.7382 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.4157 0.0722 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.9924 -0.2607 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.5691 0.0722 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.5691 0.7382 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.9924 1.0712 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.3035 0.0961 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.8596 -0.2250 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.1455 -0.2605 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.1455 -0.8971 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.6968 -1.2155 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.2482 -0.8971 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.2482 -0.2605 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.6968 0.0578 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.3982 0.6905 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0
-2.0520 0.2335 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
-1.5534 0.4274 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
-1.0337 0.4216 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
-1.4219 0.7822 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.9312 0.5993 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0
-3.2386 0.4653 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.5917 0.2072 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.2677 -0.0522 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.3163 -0.4851 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0
-3.9701 -0.9422 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
-3.4715 -0.7483 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
-2.9518 -0.7540 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
-3.3400 -0.3934 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.8493 -0.5763 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0
-5.1567 -0.7103 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.5098 -0.9684 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.1858 -1.2279 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.9924 -0.9262 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
5.1143 -1.3970 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
5.9803 -1.8970 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.5049 1.3431 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.5768 2.1650 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.8597 -1.2250 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.8597 -2.2250 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.3370 -0.6235 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.3701 -1.3306 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.6836 -0.1446 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-5.0961 0.5699 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1 2 1 0 0 0 0
2 3 2 0 0 0 0
3 4 1 0 0 0 0
4 5 2 0 0 0 0
5 6 1 0 0 0 0
6 7 1 0 0 0 0
7 8 2 0 0 0 0
8 9 1 0 0 0 0
9 4 1 0 0 0 0
2 10 1 0 0 0 0
10 11 2 0 0 0 0
11 5 1 0 0 0 0
7 12 1 0 0 0 0
12 13 2 0 0 0 0
13 14 1 0 0 0 0
14 15 2 0 0 0 0
15 16 1 0 0 0 0
16 17 2 0 0 0 0
17 12 1 0 0 0 0
18 19 1 1 0 0 0
19 20 1 1 0 0 0
21 20 1 1 0 0 0
21 22 1 0 0 0 0
22 23 1 0 0 0 0
23 18 1 0 0 0 0
18 24 1 0 0 0 0
19 25 1 0 0 0 0
20 26 1 0 0 0 0
21 1 1 0 0 0 0
27 28 1 1 0 0 0
28 29 1 1 0 0 0
30 29 1 1 0 0 0
30 31 1 0 0 0 0
31 32 1 0 0 0 0
32 27 1 0 0 0 0
27 33 1 0 0 0 0
28 34 1 0 0 0 0
29 35 1 0 0 0 0
30 25 1 0 0 0 0
6 36 2 0 0 0 0
15 37 1 0 0 0 0
37 38 1 0 0 0 0
23 39 1 0 0 0 0
39 40 1 0 0 0 0
11 41 1 0 0 0 0
41 42 1 0 0 0 0
10 43 1 0 0 0 0
43 44 1 0 0 0 0
32 45 1 0 0 0 0
45 46 1 0 0 0 0
M STY 1 5 SUP
M SLB 1 5 5
M SAL 5 2 45 46
M SBL 5 1 49
M SMT 5 CH2OH
M SVB 5 49 -4.0549 0.3403
M STY 1 4 SUP
M SLB 1 4 4
M SAL 4 2 39 40
M SBL 4 1 43
M SMT 4 CH2OH
M SVB 4 43 -2.1368 1.5159
M STY 1 3 SUP
M SLB 1 3 3
M SAL 3 2 43 44
M SBL 3 1 47
M SMT 3 OCH3
M SVB 3 47 -0.411 0.1881
M STY 1 2 SUP
M SLB 1 2 2
M SAL 2 2 41 42
M SBL 2 1 45
M SMT 2 OCH3
M SVB 2 45 0.4363 -0.3931
M STY 1 1 SUP
M SLB 1 1 1
M SAL 1 2 37 38
M SBL 1 1 41
M SMT 1 OCH3
M SVB 1 41 4.4423 -1.1034
S SKP 8
ID FLIAEAGS0008
KNApSAcK_ID C00010130
NAME 5-Methoxyafrormosin 7-O-laminaribioside
CAS_RN 68862-18-0
FORMULA C30H36O16
EXACTMASS 652.200335104
AVERAGEMASS 652.59724
SMILES O[C@@H]([C@H]4O[C@@H]([C@@H](O)5)OC([C@H](O)[C@@H]5O)CO)[C@@H](OC(CO)[C@@H]4O)Oc(c3)c(c(OC)c(c23)C(C(=CO2)c(c1)ccc(OC)c1)=O)OC
M END