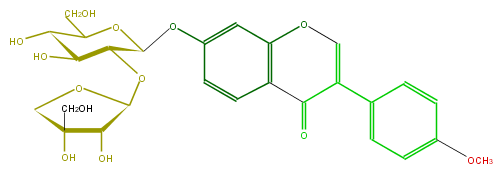
Mol:FLIA1AGS0012
From Metabolomics.JP
(Difference between revisions)
| Line 1: | Line 1: | ||
| − | + | ||
| − | + | ||
| − | Copyright: ARM project http://www.metabolome.jp/ | + | Copyright: ARM project http://www.metabolome.jp/ |
| − | 40 44 0 0 0 0 0 0 0 0999 V2000 | + | 40 44 0 0 0 0 0 0 0 0999 V2000 |
| − | -1.2946 0.9815 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.2946 0.9815 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.5738 1.3977 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.5738 1.3977 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.1467 0.9817 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.1467 0.9817 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.1467 0.1189 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.1467 0.1189 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.8939 -0.3126 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.8939 -0.3126 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.6412 0.1189 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.6412 0.1189 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.6412 0.9817 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.6412 0.9817 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.8939 1.4131 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.8939 1.4131 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.2946 0.1497 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.2946 0.1497 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.5738 -0.2662 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.5738 -0.2662 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.8939 -1.0263 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.8939 -1.0263 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.3880 -0.3123 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.3880 -0.3123 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.3880 -1.1372 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.3880 -1.1372 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.1023 -1.5496 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.1023 -1.5496 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.8168 -1.1372 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.8168 -1.1372 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.8168 -0.3123 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.8168 -0.3123 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.1023 0.1002 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.1023 0.1002 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.9688 1.3706 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.9688 1.3706 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.6964 1.2296 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.6964 1.2296 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.0687 0.6184 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.0687 0.6184 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.4004 0.8783 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.4004 0.8783 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.6304 0.8013 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.6304 0.8013 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.2242 1.3539 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.2242 1.3539 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.9069 1.1088 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.9069 1.1088 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -5.3343 1.0551 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -5.3343 1.0551 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.8112 0.7126 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.8112 0.7126 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.6655 0.2340 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.6655 0.2340 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -5.0345 -0.4784 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -5.0345 -0.4784 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.3629 -0.9375 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.3629 -0.9375 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.5444 -0.9375 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.5444 -0.9375 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.9384 -0.3786 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.9384 -0.3786 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.0485 0.0121 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.0485 0.0121 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.5444 -1.5186 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.5444 -1.5186 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.3599 -1.5103 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.3599 -1.5103 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.5708 -1.5725 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.5708 -1.5725 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 5.4966 -2.1070 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 5.4966 -2.1070 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.4120 1.6552 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.4120 1.6552 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -5.3808 2.1070 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -5.3808 2.1070 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.3641 -0.4195 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.3641 -0.4195 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -5.4966 -0.1417 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -5.4966 -0.1417 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1 2 2 0 0 0 0 | + | 1 2 2 0 0 0 0 |
| − | 2 3 1 0 0 0 0 | + | 2 3 1 0 0 0 0 |
| − | 3 4 2 0 0 0 0 | + | 3 4 2 0 0 0 0 |
| − | 4 5 1 0 0 0 0 | + | 4 5 1 0 0 0 0 |
| − | 5 6 1 0 0 0 0 | + | 5 6 1 0 0 0 0 |
| − | 6 7 2 0 0 0 0 | + | 6 7 2 0 0 0 0 |
| − | 7 8 1 0 0 0 0 | + | 7 8 1 0 0 0 0 |
| − | 8 3 1 0 0 0 0 | + | 8 3 1 0 0 0 0 |
| − | 1 9 1 0 0 0 0 | + | 1 9 1 0 0 0 0 |
| − | 9 10 2 0 0 0 0 | + | 9 10 2 0 0 0 0 |
| − | 10 4 1 0 0 0 0 | + | 10 4 1 0 0 0 0 |
| − | 5 11 2 0 0 0 0 | + | 5 11 2 0 0 0 0 |
| − | 6 12 1 0 0 0 0 | + | 6 12 1 0 0 0 0 |
| − | 12 13 2 0 0 0 0 | + | 12 13 2 0 0 0 0 |
| − | 13 14 1 0 0 0 0 | + | 13 14 1 0 0 0 0 |
| − | 14 15 2 0 0 0 0 | + | 14 15 2 0 0 0 0 |
| − | 15 16 1 0 0 0 0 | + | 15 16 1 0 0 0 0 |
| − | 16 17 2 0 0 0 0 | + | 16 17 2 0 0 0 0 |
| − | 17 12 1 0 0 0 0 | + | 17 12 1 0 0 0 0 |
| − | 1 18 1 0 0 0 0 | + | 1 18 1 0 0 0 0 |
| − | 19 20 1 1 0 0 0 | + | 19 20 1 1 0 0 0 |
| − | 20 21 1 1 0 0 0 | + | 20 21 1 1 0 0 0 |
| − | 22 21 1 1 0 0 0 | + | 22 21 1 1 0 0 0 |
| − | 22 23 1 0 0 0 0 | + | 22 23 1 0 0 0 0 |
| − | 23 24 1 0 0 0 0 | + | 23 24 1 0 0 0 0 |
| − | 24 19 1 0 0 0 0 | + | 24 19 1 0 0 0 0 |
| − | 19 25 1 0 0 0 0 | + | 19 25 1 0 0 0 0 |
| − | 20 26 1 0 0 0 0 | + | 20 26 1 0 0 0 0 |
| − | 21 27 1 0 0 0 0 | + | 21 27 1 0 0 0 0 |
| − | 18 22 1 0 0 0 0 | + | 18 22 1 0 0 0 0 |
| − | 28 29 1 1 0 0 0 | + | 28 29 1 1 0 0 0 |
| − | 29 30 1 1 0 0 0 | + | 29 30 1 1 0 0 0 |
| − | 31 30 1 1 0 0 0 | + | 31 30 1 1 0 0 0 |
| − | 31 32 1 0 0 0 0 | + | 31 32 1 0 0 0 0 |
| − | 32 28 1 0 0 0 0 | + | 32 28 1 0 0 0 0 |
| − | 30 33 1 0 0 0 0 | + | 30 33 1 0 0 0 0 |
| − | 31 27 1 0 0 0 0 | + | 31 27 1 0 0 0 0 |
| − | 29 34 1 0 0 0 0 | + | 29 34 1 0 0 0 0 |
| − | 35 36 1 0 0 0 0 | + | 35 36 1 0 0 0 0 |
| − | 15 35 1 0 0 0 0 | + | 15 35 1 0 0 0 0 |
| − | 37 38 1 0 0 0 0 | + | 37 38 1 0 0 0 0 |
| − | 24 37 1 0 0 0 0 | + | 24 37 1 0 0 0 0 |
| − | 39 40 1 0 0 0 0 | + | 39 40 1 0 0 0 0 |
| − | 29 39 1 0 0 0 0 | + | 29 39 1 0 0 0 0 |
| − | M STY 1 1 SUP | + | M STY 1 1 SUP |
| − | M SLB 1 1 1 | + | M SLB 1 1 1 |
| − | M SAL 1 2 35 36 | + | M SAL 1 2 35 36 |
| − | M SBL 1 1 40 | + | M SBL 1 1 40 |
| − | M SMT 1 OCH3 | + | M SMT 1 OCH3 |
| − | M SBV 1 40 -0.7540 0.4353 | + | M SBV 1 40 -0.7540 0.4353 |
| − | M STY 1 2 SUP | + | M STY 1 2 SUP |
| − | M SLB 1 2 2 | + | M SLB 1 2 2 |
| − | M SAL 2 2 37 38 | + | M SAL 2 2 37 38 |
| − | M SBL 2 1 42 | + | M SBL 2 1 42 |
| − | M SMT 2 ^ CH2OH | + | M SMT 2 ^ CH2OH |
| − | M SBV 2 42 0.5051 -0.5464 | + | M SBV 2 42 0.5051 -0.5464 |
| − | M STY 1 3 SUP | + | M STY 1 3 SUP |
| − | M SLB 1 3 3 | + | M SLB 1 3 3 |
| − | M SAL 3 2 39 40 | + | M SAL 3 2 39 40 |
| − | M SBL 3 1 44 | + | M SBL 3 1 44 |
| − | M SMT 3 CH2OH | + | M SMT 3 CH2OH |
| − | M SBV 3 44 0.0012 -0.5181 | + | M SBV 3 44 0.0012 -0.5181 |
| − | S SKP 5 | + | S SKP 5 |
| − | ID FLIA1AGS0012 | + | ID FLIA1AGS0012 |
| − | FORMULA C27H30O13 | + | FORMULA C27H30O13 |
| − | EXACTMASS 562.168641046 | + | EXACTMASS 562.168641046 |
| − | AVERAGEMASS 562.5193 | + | AVERAGEMASS 562.5193 |
| − | SMILES O(C(C(OC(O5)C(O)C(O)(C5)CO)4)OC(CO)C(C4O)O)c(c3)cc(c1c3)OC=C(c(c2)ccc(OC)c2)C1=O | + | SMILES O(C(C(OC(O5)C(O)C(O)(C5)CO)4)OC(CO)C(C4O)O)c(c3)cc(c1c3)OC=C(c(c2)ccc(OC)c2)C1=O |
M END | M END | ||
| − | |||
Latest revision as of 09:00, 14 March 2009
Copyright: ARM project http://www.metabolome.jp/
40 44 0 0 0 0 0 0 0 0999 V2000
-1.2946 0.9815 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.5738 1.3977 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.1467 0.9817 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.1467 0.1189 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.8939 -0.3126 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.6412 0.1189 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.6412 0.9817 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.8939 1.4131 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.2946 0.1497 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.5738 -0.2662 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.8939 -1.0263 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.3880 -0.3123 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.3880 -1.1372 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.1023 -1.5496 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.8168 -1.1372 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.8168 -0.3123 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.1023 0.1002 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.9688 1.3706 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.6964 1.2296 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.0687 0.6184 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.4004 0.8783 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.6304 0.8013 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.2242 1.3539 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.9069 1.1088 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-5.3343 1.0551 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.8112 0.7126 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.6655 0.2340 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-5.0345 -0.4784 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.3629 -0.9375 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.5444 -0.9375 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.9384 -0.3786 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.0485 0.0121 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.5444 -1.5186 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.3599 -1.5103 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
4.5708 -1.5725 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
5.4966 -2.1070 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.4120 1.6552 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-5.3808 2.1070 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.3641 -0.4195 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-5.4966 -0.1417 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1 2 2 0 0 0 0
2 3 1 0 0 0 0
3 4 2 0 0 0 0
4 5 1 0 0 0 0
5 6 1 0 0 0 0
6 7 2 0 0 0 0
7 8 1 0 0 0 0
8 3 1 0 0 0 0
1 9 1 0 0 0 0
9 10 2 0 0 0 0
10 4 1 0 0 0 0
5 11 2 0 0 0 0
6 12 1 0 0 0 0
12 13 2 0 0 0 0
13 14 1 0 0 0 0
14 15 2 0 0 0 0
15 16 1 0 0 0 0
16 17 2 0 0 0 0
17 12 1 0 0 0 0
1 18 1 0 0 0 0
19 20 1 1 0 0 0
20 21 1 1 0 0 0
22 21 1 1 0 0 0
22 23 1 0 0 0 0
23 24 1 0 0 0 0
24 19 1 0 0 0 0
19 25 1 0 0 0 0
20 26 1 0 0 0 0
21 27 1 0 0 0 0
18 22 1 0 0 0 0
28 29 1 1 0 0 0
29 30 1 1 0 0 0
31 30 1 1 0 0 0
31 32 1 0 0 0 0
32 28 1 0 0 0 0
30 33 1 0 0 0 0
31 27 1 0 0 0 0
29 34 1 0 0 0 0
35 36 1 0 0 0 0
15 35 1 0 0 0 0
37 38 1 0 0 0 0
24 37 1 0 0 0 0
39 40 1 0 0 0 0
29 39 1 0 0 0 0
M STY 1 1 SUP
M SLB 1 1 1
M SAL 1 2 35 36
M SBL 1 1 40
M SMT 1 OCH3
M SBV 1 40 -0.7540 0.4353
M STY 1 2 SUP
M SLB 1 2 2
M SAL 2 2 37 38
M SBL 2 1 42
M SMT 2 ^ CH2OH
M SBV 2 42 0.5051 -0.5464
M STY 1 3 SUP
M SLB 1 3 3
M SAL 3 2 39 40
M SBL 3 1 44
M SMT 3 CH2OH
M SBV 3 44 0.0012 -0.5181
S SKP 5
ID FLIA1AGS0012
FORMULA C27H30O13
EXACTMASS 562.168641046
AVERAGEMASS 562.5193
SMILES O(C(C(OC(O5)C(O)C(O)(C5)CO)4)OC(CO)C(C4O)O)c(c3)cc(c1c3)OC=C(c(c2)ccc(OC)c2)C1=O
M END