Mol:FL7AAIGL0006
From Metabolomics.JP
(Difference between revisions)
| Line 1: | Line 1: | ||
| − | + | ||
| − | + | ||
| − | Copyright: ARM project http://www.metabolome.jp/ | + | Copyright: ARM project http://www.metabolome.jp/ |
| − | 46 50 0 0 0 0 0 0 0 0999 V2000 | + | 46 50 0 0 0 0 0 0 0 0999 V2000 |
| − | -1.2891 0.0453 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.2891 0.0453 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.2891 -0.5971 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.2891 -0.5971 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.7328 -0.9182 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.7328 -0.9182 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.1765 -0.5971 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.1765 -0.5971 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.1765 0.0453 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.1765 0.0453 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.7328 0.3665 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.7328 0.3665 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.3798 -0.9182 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.3798 -0.9182 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.9361 -0.5971 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.9361 -0.5971 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.9361 0.0453 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.9361 0.0453 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.3798 0.3665 0.0000 O 0 3 0 0 0 0 0 0 0 0 0 0 | + | 0.3798 0.3665 0.0000 O 0 3 0 0 0 0 0 0 0 0 0 0 |
| − | 1.4922 0.3664 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.4922 0.3664 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.0592 0.0390 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.0592 0.0390 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.6261 0.3664 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.6261 0.3664 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.6261 1.0210 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.6261 1.0210 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.0592 1.3484 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.0592 1.3484 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.4922 1.0210 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.4922 1.0210 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.8452 0.3664 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.8452 0.3664 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.4921 1.5210 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.4921 1.5210 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.7328 -1.5604 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.7328 -1.5604 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.3522 -1.3178 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.3522 -1.3178 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.5096 -1.0177 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 | + | 2.5096 -1.0177 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 |
| − | 2.2088 -1.5387 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | 2.2088 -1.5387 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | 2.7873 -1.3734 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.7873 -1.3734 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.3692 -1.5387 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | 3.3692 -1.5387 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | 3.6701 -1.0177 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | 3.6701 -1.0177 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | 3.0916 -1.1830 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 | + | 3.0916 -1.1830 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 |
| − | 1.9509 -1.1674 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.9509 -1.1674 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.1331 -0.7084 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.1331 -0.7084 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.1042 -1.2683 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.1042 -1.2683 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.9228 0.1867 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0 | + | -3.9228 0.1867 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0 |
| − | -3.5517 -0.3033 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | -3.5517 -0.3033 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | -3.0172 -0.0954 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | -3.0172 -0.0954 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | -2.4062 -0.2435 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | -2.4062 -0.2435 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | -2.8762 0.2850 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.8762 0.2850 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.3438 0.0382 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 | + | -3.3438 0.0382 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 |
| − | -4.5138 0.2384 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.5138 0.2384 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.9021 -0.7645 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.9021 -0.7645 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.7109 -0.6095 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.7109 -0.6095 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.5688 -1.8311 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.5688 -1.8311 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.5688 -2.6561 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.5688 -2.6561 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.6261 0.3664 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.6261 0.3664 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.6261 0.3664 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.6261 0.3664 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.3426 1.9245 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.3426 1.9245 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.7840 2.8218 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.7840 2.8218 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.0363 0.5806 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.0363 0.5806 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.3184 1.3557 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.3184 1.3557 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1 2 1 0 0 0 0 | + | 1 2 1 0 0 0 0 |
| − | 2 3 2 0 0 0 0 | + | 2 3 2 0 0 0 0 |
| − | 3 4 1 0 0 0 0 | + | 3 4 1 0 0 0 0 |
| − | 4 5 2 0 0 0 0 | + | 4 5 2 0 0 0 0 |
| − | 5 6 1 0 0 0 0 | + | 5 6 1 0 0 0 0 |
| − | 6 1 2 0 0 0 0 | + | 6 1 2 0 0 0 0 |
| − | 4 7 1 0 0 0 0 | + | 4 7 1 0 0 0 0 |
| − | 7 8 2 0 0 0 0 | + | 7 8 2 0 0 0 0 |
| − | 8 9 1 0 0 0 0 | + | 8 9 1 0 0 0 0 |
| − | 9 10 2 0 0 0 0 | + | 9 10 2 0 0 0 0 |
| − | 10 5 1 0 0 0 0 | + | 10 5 1 0 0 0 0 |
| − | 9 11 1 0 0 0 0 | + | 9 11 1 0 0 0 0 |
| − | 11 12 2 0 0 0 0 | + | 11 12 2 0 0 0 0 |
| − | 12 13 1 0 0 0 0 | + | 12 13 1 0 0 0 0 |
| − | 13 14 2 0 0 0 0 | + | 13 14 2 0 0 0 0 |
| − | 14 15 1 0 0 0 0 | + | 14 15 1 0 0 0 0 |
| − | 15 16 2 0 0 0 0 | + | 15 16 2 0 0 0 0 |
| − | 16 11 1 0 0 0 0 | + | 16 11 1 0 0 0 0 |
| − | 1 17 1 0 0 0 0 | + | 1 17 1 0 0 0 0 |
| − | 14 18 1 0 0 0 0 | + | 14 18 1 0 0 0 0 |
| − | 3 19 1 0 0 0 0 | + | 3 19 1 0 0 0 0 |
| − | 20 8 1 0 0 0 0 | + | 20 8 1 0 0 0 0 |
| − | 21 22 1 0 0 0 0 | + | 21 22 1 0 0 0 0 |
| − | 22 23 1 1 0 0 0 | + | 22 23 1 1 0 0 0 |
| − | 23 24 1 1 0 0 0 | + | 23 24 1 1 0 0 0 |
| − | 25 24 1 1 0 0 0 | + | 25 24 1 1 0 0 0 |
| − | 25 26 1 0 0 0 0 | + | 25 26 1 0 0 0 0 |
| − | 26 21 1 0 0 0 0 | + | 26 21 1 0 0 0 0 |
| − | 21 27 1 0 0 0 0 | + | 21 27 1 0 0 0 0 |
| − | 26 28 1 0 0 0 0 | + | 26 28 1 0 0 0 0 |
| − | 25 29 1 0 0 0 0 | + | 25 29 1 0 0 0 0 |
| − | 22 20 1 0 0 0 0 | + | 22 20 1 0 0 0 0 |
| − | 30 31 1 1 0 0 0 | + | 30 31 1 1 0 0 0 |
| − | 31 32 1 1 0 0 0 | + | 31 32 1 1 0 0 0 |
| − | 33 32 1 1 0 0 0 | + | 33 32 1 1 0 0 0 |
| − | 33 34 1 0 0 0 0 | + | 33 34 1 0 0 0 0 |
| − | 34 35 1 0 0 0 0 | + | 34 35 1 0 0 0 0 |
| − | 35 30 1 0 0 0 0 | + | 35 30 1 0 0 0 0 |
| − | 30 36 1 0 0 0 0 | + | 30 36 1 0 0 0 0 |
| − | 31 37 1 0 0 0 0 | + | 31 37 1 0 0 0 0 |
| − | 32 38 1 0 0 0 0 | + | 32 38 1 0 0 0 0 |
| − | 33 17 1 0 0 0 0 | + | 33 17 1 0 0 0 0 |
| − | 24 39 1 0 0 0 0 | + | 24 39 1 0 0 0 0 |
| − | 39 40 1 0 0 0 0 | + | 39 40 1 0 0 0 0 |
| − | 13 41 1 0 0 0 0 | + | 13 41 1 0 0 0 0 |
| − | 41 42 1 0 0 0 0 | + | 41 42 1 0 0 0 0 |
| − | 15 43 1 0 0 0 0 | + | 15 43 1 0 0 0 0 |
| − | 43 44 1 0 0 0 0 | + | 43 44 1 0 0 0 0 |
| − | 35 45 1 0 0 0 0 | + | 35 45 1 0 0 0 0 |
| − | 45 46 1 0 0 0 0 | + | 45 46 1 0 0 0 0 |
| − | M STY 1 4 SUP | + | M STY 1 4 SUP |
| − | M SLB 1 4 4 | + | M SLB 1 4 4 |
| − | M SAL 4 2 45 46 | + | M SAL 4 2 45 46 |
| − | M SBL 4 1 49 | + | M SBL 4 1 49 |
| − | M SMT 4 CH2OH | + | M SMT 4 CH2OH |
| − | M SVB 4 49 -3.7422 0.8224 | + | M SVB 4 49 -3.7422 0.8224 |
| − | M STY 1 3 SUP | + | M STY 1 3 SUP |
| − | M SLB 1 3 3 | + | M SLB 1 3 3 |
| − | M SAL 3 2 39 40 | + | M SAL 3 2 39 40 |
| − | M SBL 3 1 43 | + | M SBL 3 1 43 |
| − | M SMT 3 CH2OH | + | M SMT 3 CH2OH |
| − | M SVB 3 43 3.7222 -1.512 | + | M SVB 3 43 3.7222 -1.512 |
| − | M STY 1 2 SUP | + | M STY 1 2 SUP |
| − | M SLB 1 2 2 | + | M SLB 1 2 2 |
| − | M SAL 2 2 43 44 | + | M SAL 2 2 43 44 |
| − | M SBL 2 1 47 | + | M SBL 2 1 47 |
| − | M SMT 2 OCH3 | + | M SMT 2 OCH3 |
| − | M SVB 2 47 2.3426 1.9245 | + | M SVB 2 47 2.3426 1.9245 |
| − | M STY 1 1 SUP | + | M STY 1 1 SUP |
| − | M SLB 1 1 1 | + | M SLB 1 1 1 |
| − | M SAL 1 2 41 42 | + | M SAL 1 2 41 42 |
| − | M SBL 1 1 45 | + | M SBL 1 1 45 |
| − | M SMT 1 OCH3 | + | M SMT 1 OCH3 |
| − | M SVB 1 45 2.8357 0.1601 | + | M SVB 1 45 2.8357 0.1601 |
| − | S SKP 8 | + | S SKP 8 |
| − | ID FL7AAIGL0006 | + | ID FL7AAIGL0006 |
| − | KNApSAcK_ID C00006742 | + | KNApSAcK_ID C00006742 |
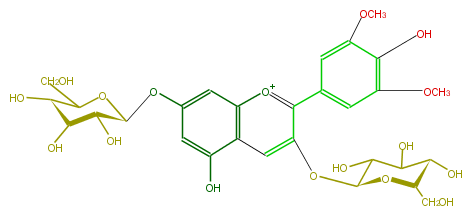
| − | NAME Malvidin 3,7-diglucoside | + | NAME Malvidin 3,7-diglucoside |
| − | CAS_RN 96407-95-3 | + | CAS_RN 96407-95-3 |
| − | FORMULA C29H35O17 | + | FORMULA C29H35O17 |
| − | EXACTMASS 655.187424694 | + | EXACTMASS 655.187424694 |
| − | AVERAGEMASS 655.578 | + | AVERAGEMASS 655.578 |
| − | SMILES [o+1](c(c(c5)cc(OC)c(O)c(OC)5)1)c(c3)c(c(cc3O[C@H](O4)[C@H]([C@@H](O)[C@@H](O)C4CO)O)O)cc1O[C@@H](C(O)2)O[C@@H]([C@H](O)C2O)CO | + | SMILES [o+1](c(c(c5)cc(OC)c(O)c(OC)5)1)c(c3)c(c(cc3O[C@H](O4)[C@H]([C@@H](O)[C@@H](O)C4CO)O)O)cc1O[C@@H](C(O)2)O[C@@H]([C@H](O)C2O)CO |
M END | M END | ||
| − | |||
Latest revision as of 09:00, 14 March 2009
Copyright: ARM project http://www.metabolome.jp/
46 50 0 0 0 0 0 0 0 0999 V2000
-1.2891 0.0453 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.2891 -0.5971 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.7328 -0.9182 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.1765 -0.5971 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.1765 0.0453 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.7328 0.3665 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.3798 -0.9182 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.9361 -0.5971 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.9361 0.0453 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.3798 0.3665 0.0000 O 0 3 0 0 0 0 0 0 0 0 0 0
1.4922 0.3664 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.0592 0.0390 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.6261 0.3664 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.6261 1.0210 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.0592 1.3484 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.4922 1.0210 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.8452 0.3664 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.4921 1.5210 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.7328 -1.5604 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.3522 -1.3178 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.5096 -1.0177 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0
2.2088 -1.5387 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
2.7873 -1.3734 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.3692 -1.5387 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
3.6701 -1.0177 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
3.0916 -1.1830 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0
1.9509 -1.1674 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.1331 -0.7084 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
4.1042 -1.2683 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.9228 0.1867 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0
-3.5517 -0.3033 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
-3.0172 -0.0954 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
-2.4062 -0.2435 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
-2.8762 0.2850 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.3438 0.0382 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0
-4.5138 0.2384 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.9021 -0.7645 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.7109 -0.6095 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.5688 -1.8311 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.5688 -2.6561 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.6261 0.3664 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
4.6261 0.3664 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.3426 1.9245 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.7840 2.8218 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.0363 0.5806 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.3184 1.3557 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1 2 1 0 0 0 0
2 3 2 0 0 0 0
3 4 1 0 0 0 0
4 5 2 0 0 0 0
5 6 1 0 0 0 0
6 1 2 0 0 0 0
4 7 1 0 0 0 0
7 8 2 0 0 0 0
8 9 1 0 0 0 0
9 10 2 0 0 0 0
10 5 1 0 0 0 0
9 11 1 0 0 0 0
11 12 2 0 0 0 0
12 13 1 0 0 0 0
13 14 2 0 0 0 0
14 15 1 0 0 0 0
15 16 2 0 0 0 0
16 11 1 0 0 0 0
1 17 1 0 0 0 0
14 18 1 0 0 0 0
3 19 1 0 0 0 0
20 8 1 0 0 0 0
21 22 1 0 0 0 0
22 23 1 1 0 0 0
23 24 1 1 0 0 0
25 24 1 1 0 0 0
25 26 1 0 0 0 0
26 21 1 0 0 0 0
21 27 1 0 0 0 0
26 28 1 0 0 0 0
25 29 1 0 0 0 0
22 20 1 0 0 0 0
30 31 1 1 0 0 0
31 32 1 1 0 0 0
33 32 1 1 0 0 0
33 34 1 0 0 0 0
34 35 1 0 0 0 0
35 30 1 0 0 0 0
30 36 1 0 0 0 0
31 37 1 0 0 0 0
32 38 1 0 0 0 0
33 17 1 0 0 0 0
24 39 1 0 0 0 0
39 40 1 0 0 0 0
13 41 1 0 0 0 0
41 42 1 0 0 0 0
15 43 1 0 0 0 0
43 44 1 0 0 0 0
35 45 1 0 0 0 0
45 46 1 0 0 0 0
M STY 1 4 SUP
M SLB 1 4 4
M SAL 4 2 45 46
M SBL 4 1 49
M SMT 4 CH2OH
M SVB 4 49 -3.7422 0.8224
M STY 1 3 SUP
M SLB 1 3 3
M SAL 3 2 39 40
M SBL 3 1 43
M SMT 3 CH2OH
M SVB 3 43 3.7222 -1.512
M STY 1 2 SUP
M SLB 1 2 2
M SAL 2 2 43 44
M SBL 2 1 47
M SMT 2 OCH3
M SVB 2 47 2.3426 1.9245
M STY 1 1 SUP
M SLB 1 1 1
M SAL 1 2 41 42
M SBL 1 1 45
M SMT 1 OCH3
M SVB 1 45 2.8357 0.1601
S SKP 8
ID FL7AAIGL0006
KNApSAcK_ID C00006742
NAME Malvidin 3,7-diglucoside
CAS_RN 96407-95-3
FORMULA C29H35O17
EXACTMASS 655.187424694
AVERAGEMASS 655.578
SMILES [o+1](c(c(c5)cc(OC)c(O)c(OC)5)1)c(c3)c(c(cc3O[C@H](O4)[C@H]([C@@H](O)[C@@H](O)C4CO)O)O)cc1O[C@@H](C(O)2)O[C@@H]([C@H](O)C2O)CO
M END