Mol:FL7AAAGL0033
From Metabolomics.JP
(Difference between revisions)
| Line 1: | Line 1: | ||
| − | + | ||
| − | + | ||
| − | Copyright: ARM project http://www.metabolome.jp/ | + | Copyright: ARM project http://www.metabolome.jp/ |
| − | 56 61 0 0 0 0 0 0 0 0999 V2000 | + | 56 61 0 0 0 0 0 0 0 0999 V2000 |
| − | -1.4431 1.2521 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.4431 1.2521 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.4431 0.6097 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.4431 0.6097 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.8868 0.2885 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.8868 0.2885 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.3305 0.6097 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.3305 0.6097 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.3305 1.2521 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.3305 1.2521 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.8868 1.5732 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.8868 1.5732 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.2258 0.2885 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.2258 0.2885 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.7821 0.6097 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.7821 0.6097 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.7821 1.2521 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.7821 1.2521 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.2258 1.5732 0.0000 O 0 3 0 0 0 0 0 0 0 0 0 0 | + | 0.2258 1.5732 0.0000 O 0 3 0 0 0 0 0 0 0 0 0 0 |
| − | 1.3382 1.5731 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.3382 1.5731 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.9052 1.2458 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.9052 1.2458 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.4722 1.5731 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.4722 1.5731 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.4722 2.2278 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.4722 2.2278 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.9052 2.5551 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.9052 2.5551 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.3382 2.2278 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.3382 2.2278 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.9992 1.5731 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.9992 1.5731 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.0390 2.5551 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.0390 2.5551 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.8868 -0.3536 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.8868 -0.3536 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.1982 -0.1110 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.1982 -0.1110 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.3556 0.1891 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 | + | 2.3556 0.1891 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 |
| − | 2.0548 -0.3320 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | 2.0548 -0.3320 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | 2.6333 -0.1666 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.6333 -0.1666 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.2153 -0.3320 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | 3.2153 -0.3320 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | 3.5162 0.1891 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | 3.5162 0.1891 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | 2.9376 0.0238 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 | + | 2.9376 0.0238 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 |
| − | 1.7970 0.0394 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.7970 0.0394 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.9792 0.4983 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.9792 0.4983 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.8706 0.5435 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.8706 0.5435 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.7668 -0.3320 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.7668 -0.3320 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.2105 -0.5247 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0 | + | -3.2105 -0.5247 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0 |
| − | -2.8394 -1.0146 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | -2.8394 -1.0146 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | -2.3049 -0.8068 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | -2.3049 -0.8068 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | -1.6939 -0.9548 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | -1.6939 -0.9548 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | -2.1639 -0.4264 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.1639 -0.4264 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.6315 -0.6732 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 | + | -2.6315 -0.6732 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 |
| − | -3.7680 -0.7275 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.7680 -0.7275 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.9620 -1.5807 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.9620 -1.5807 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.9986 -1.3209 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.9986 -1.3209 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.4893 -0.1593 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.4893 -0.1593 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.9574 -1.0432 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.9574 -1.0432 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.6680 -1.5444 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.6680 -1.5444 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.1528 -1.5444 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.1528 -1.5444 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.8585 -2.0542 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.8585 -2.0542 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.3692 -2.0542 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.3692 -2.0542 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.0800 -1.5533 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.0800 -1.5533 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.5016 -1.5533 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.5016 -1.5533 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.2124 -2.0542 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.2124 -2.0542 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.5016 -2.5551 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.5016 -2.5551 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.0800 -2.5551 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.0800 -2.5551 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.6349 -2.0542 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.6349 -2.0542 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.9637 -2.0565 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.9637 -2.0565 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.8204 0.4774 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.8204 0.4774 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.2419 0.8312 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.2419 0.8312 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.7517 0.6457 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.7517 0.6457 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.1464 1.3728 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.1464 1.3728 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1 2 1 0 0 0 0 | + | 1 2 1 0 0 0 0 |
| − | 2 3 2 0 0 0 0 | + | 2 3 2 0 0 0 0 |
| − | 3 4 1 0 0 0 0 | + | 3 4 1 0 0 0 0 |
| − | 4 5 2 0 0 0 0 | + | 4 5 2 0 0 0 0 |
| − | 5 6 1 0 0 0 0 | + | 5 6 1 0 0 0 0 |
| − | 6 1 2 0 0 0 0 | + | 6 1 2 0 0 0 0 |
| − | 4 7 1 0 0 0 0 | + | 4 7 1 0 0 0 0 |
| − | 7 8 2 0 0 0 0 | + | 7 8 2 0 0 0 0 |
| − | 8 9 1 0 0 0 0 | + | 8 9 1 0 0 0 0 |
| − | 9 10 2 0 0 0 0 | + | 9 10 2 0 0 0 0 |
| − | 10 5 1 0 0 0 0 | + | 10 5 1 0 0 0 0 |
| − | 9 11 1 0 0 0 0 | + | 9 11 1 0 0 0 0 |
| − | 11 12 2 0 0 0 0 | + | 11 12 2 0 0 0 0 |
| − | 12 13 1 0 0 0 0 | + | 12 13 1 0 0 0 0 |
| − | 13 14 2 0 0 0 0 | + | 13 14 2 0 0 0 0 |
| − | 14 15 1 0 0 0 0 | + | 14 15 1 0 0 0 0 |
| − | 15 16 2 0 0 0 0 | + | 15 16 2 0 0 0 0 |
| − | 16 11 1 0 0 0 0 | + | 16 11 1 0 0 0 0 |
| − | 1 17 1 0 0 0 0 | + | 1 17 1 0 0 0 0 |
| − | 14 18 1 0 0 0 0 | + | 14 18 1 0 0 0 0 |
| − | 3 19 1 0 0 0 0 | + | 3 19 1 0 0 0 0 |
| − | 20 8 1 0 0 0 0 | + | 20 8 1 0 0 0 0 |
| − | 21 22 1 0 0 0 0 | + | 21 22 1 0 0 0 0 |
| − | 22 23 1 1 0 0 0 | + | 22 23 1 1 0 0 0 |
| − | 23 24 1 1 0 0 0 | + | 23 24 1 1 0 0 0 |
| − | 25 24 1 1 0 0 0 | + | 25 24 1 1 0 0 0 |
| − | 25 26 1 0 0 0 0 | + | 25 26 1 0 0 0 0 |
| − | 26 21 1 0 0 0 0 | + | 26 21 1 0 0 0 0 |
| − | 21 27 1 0 0 0 0 | + | 21 27 1 0 0 0 0 |
| − | 26 28 1 0 0 0 0 | + | 26 28 1 0 0 0 0 |
| − | 25 29 1 0 0 0 0 | + | 25 29 1 0 0 0 0 |
| − | 24 30 1 0 0 0 0 | + | 24 30 1 0 0 0 0 |
| − | 22 20 1 0 0 0 0 | + | 22 20 1 0 0 0 0 |
| − | 31 32 1 1 0 0 0 | + | 31 32 1 1 0 0 0 |
| − | 32 33 1 1 0 0 0 | + | 32 33 1 1 0 0 0 |
| − | 34 33 1 1 0 0 0 | + | 34 33 1 1 0 0 0 |
| − | 34 35 1 0 0 0 0 | + | 34 35 1 0 0 0 0 |
| − | 35 36 1 0 0 0 0 | + | 35 36 1 0 0 0 0 |
| − | 36 31 1 0 0 0 0 | + | 36 31 1 0 0 0 0 |
| − | 31 37 1 0 0 0 0 | + | 31 37 1 0 0 0 0 |
| − | 32 38 1 0 0 0 0 | + | 32 38 1 0 0 0 0 |
| − | 33 39 1 0 0 0 0 | + | 33 39 1 0 0 0 0 |
| − | 36 40 1 0 0 0 0 | + | 36 40 1 0 0 0 0 |
| − | 34 19 1 0 0 0 0 | + | 34 19 1 0 0 0 0 |
| − | 30 41 1 0 0 0 0 | + | 30 41 1 0 0 0 0 |
| − | 41 42 1 0 0 0 0 | + | 41 42 1 0 0 0 0 |
| − | 42 43 1 0 0 0 0 | + | 42 43 1 0 0 0 0 |
| − | 43 44 2 0 0 0 0 | + | 43 44 2 0 0 0 0 |
| − | 44 45 1 0 0 0 0 | + | 44 45 1 0 0 0 0 |
| − | 45 46 2 0 0 0 0 | + | 45 46 2 0 0 0 0 |
| − | 46 47 1 0 0 0 0 | + | 46 47 1 0 0 0 0 |
| − | 47 48 2 0 0 0 0 | + | 47 48 2 0 0 0 0 |
| − | 48 49 1 0 0 0 0 | + | 48 49 1 0 0 0 0 |
| − | 49 50 2 0 0 0 0 | + | 49 50 2 0 0 0 0 |
| − | 50 45 1 0 0 0 0 | + | 50 45 1 0 0 0 0 |
| − | 48 51 1 0 0 0 0 | + | 48 51 1 0 0 0 0 |
| − | 42 52 2 0 0 0 0 | + | 42 52 2 0 0 0 0 |
| − | 40 53 1 0 0 0 0 | + | 40 53 1 0 0 0 0 |
| − | 53 54 1 0 0 0 0 | + | 53 54 1 0 0 0 0 |
| − | 54 55 1 0 0 0 0 | + | 54 55 1 0 0 0 0 |
| − | 54 56 2 0 0 0 0 | + | 54 56 2 0 0 0 0 |
| − | S SKP 8 | + | S SKP 8 |
| − | ID FL7AAAGL0033 | + | ID FL7AAAGL0033 |
| − | KNApSAcK_ID C00006772 | + | KNApSAcK_ID C00006772 |
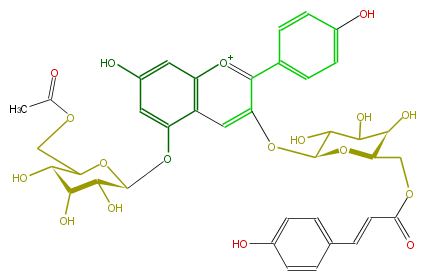
| − | NAME Pelargonidin 3-(6''-p-coumarylglucoside)-5-(6'''-acetylglucoside) | + | NAME Pelargonidin 3-(6''-p-coumarylglucoside)-5-(6'''-acetylglucoside) |
| − | CAS_RN 168647-44-7 | + | CAS_RN 168647-44-7 |
| − | FORMULA C38H39O18 | + | FORMULA C38H39O18 |
| − | EXACTMASS 783.213639444 | + | EXACTMASS 783.213639444 |
| − | AVERAGEMASS 783.70546 | + | AVERAGEMASS 783.70546 |
| − | SMILES O(c(c3c(c6)ccc(O)c6)cc(c4O[C@H](O5)[C@H]([C@@H](O)[C@H](C5COC(C)=O)O)O)c(cc(c4)O)[o+1]3)[C@H](O1)C(O)C([C@@H](O)[C@H]1COC(C=Cc(c2)ccc(O)c2)=O)O | + | SMILES O(c(c3c(c6)ccc(O)c6)cc(c4O[C@H](O5)[C@H]([C@@H](O)[C@H](C5COC(C)=O)O)O)c(cc(c4)O)[o+1]3)[C@H](O1)C(O)C([C@@H](O)[C@H]1COC(C=Cc(c2)ccc(O)c2)=O)O |
M END | M END | ||
| − | |||
Latest revision as of 09:00, 14 March 2009
Copyright: ARM project http://www.metabolome.jp/
56 61 0 0 0 0 0 0 0 0999 V2000
-1.4431 1.2521 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.4431 0.6097 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.8868 0.2885 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.3305 0.6097 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.3305 1.2521 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.8868 1.5732 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.2258 0.2885 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.7821 0.6097 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.7821 1.2521 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.2258 1.5732 0.0000 O 0 3 0 0 0 0 0 0 0 0 0 0
1.3382 1.5731 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.9052 1.2458 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.4722 1.5731 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.4722 2.2278 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.9052 2.5551 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.3382 2.2278 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.9992 1.5731 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.0390 2.5551 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.8868 -0.3536 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.1982 -0.1110 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.3556 0.1891 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0
2.0548 -0.3320 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
2.6333 -0.1666 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.2153 -0.3320 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
3.5162 0.1891 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
2.9376 0.0238 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0
1.7970 0.0394 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.9792 0.4983 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.8706 0.5435 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.7668 -0.3320 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.2105 -0.5247 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0
-2.8394 -1.0146 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
-2.3049 -0.8068 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
-1.6939 -0.9548 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
-2.1639 -0.4264 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.6315 -0.6732 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0
-3.7680 -0.7275 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.9620 -1.5807 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.9986 -1.3209 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.4893 -0.1593 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.9574 -1.0432 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.6680 -1.5444 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.1528 -1.5444 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.8585 -2.0542 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.3692 -2.0542 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.0800 -1.5533 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.5016 -1.5533 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.2124 -2.0542 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.5016 -2.5551 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.0800 -2.5551 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.6349 -2.0542 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.9637 -2.0565 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.8204 0.4774 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.2419 0.8312 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.7517 0.6457 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.1464 1.3728 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1 2 1 0 0 0 0
2 3 2 0 0 0 0
3 4 1 0 0 0 0
4 5 2 0 0 0 0
5 6 1 0 0 0 0
6 1 2 0 0 0 0
4 7 1 0 0 0 0
7 8 2 0 0 0 0
8 9 1 0 0 0 0
9 10 2 0 0 0 0
10 5 1 0 0 0 0
9 11 1 0 0 0 0
11 12 2 0 0 0 0
12 13 1 0 0 0 0
13 14 2 0 0 0 0
14 15 1 0 0 0 0
15 16 2 0 0 0 0
16 11 1 0 0 0 0
1 17 1 0 0 0 0
14 18 1 0 0 0 0
3 19 1 0 0 0 0
20 8 1 0 0 0 0
21 22 1 0 0 0 0
22 23 1 1 0 0 0
23 24 1 1 0 0 0
25 24 1 1 0 0 0
25 26 1 0 0 0 0
26 21 1 0 0 0 0
21 27 1 0 0 0 0
26 28 1 0 0 0 0
25 29 1 0 0 0 0
24 30 1 0 0 0 0
22 20 1 0 0 0 0
31 32 1 1 0 0 0
32 33 1 1 0 0 0
34 33 1 1 0 0 0
34 35 1 0 0 0 0
35 36 1 0 0 0 0
36 31 1 0 0 0 0
31 37 1 0 0 0 0
32 38 1 0 0 0 0
33 39 1 0 0 0 0
36 40 1 0 0 0 0
34 19 1 0 0 0 0
30 41 1 0 0 0 0
41 42 1 0 0 0 0
42 43 1 0 0 0 0
43 44 2 0 0 0 0
44 45 1 0 0 0 0
45 46 2 0 0 0 0
46 47 1 0 0 0 0
47 48 2 0 0 0 0
48 49 1 0 0 0 0
49 50 2 0 0 0 0
50 45 1 0 0 0 0
48 51 1 0 0 0 0
42 52 2 0 0 0 0
40 53 1 0 0 0 0
53 54 1 0 0 0 0
54 55 1 0 0 0 0
54 56 2 0 0 0 0
S SKP 8
ID FL7AAAGL0033
KNApSAcK_ID C00006772
NAME Pelargonidin 3-(6''-p-coumarylglucoside)-5-(6'''-acetylglucoside)
CAS_RN 168647-44-7
FORMULA C38H39O18
EXACTMASS 783.213639444
AVERAGEMASS 783.70546
SMILES O(c(c3c(c6)ccc(O)c6)cc(c4O[C@H](O5)[C@H]([C@@H](O)[C@H](C5COC(C)=O)O)O)c(cc(c4)O)[o+1]3)[C@H](O1)C(O)C([C@@H](O)[C@H]1COC(C=Cc(c2)ccc(O)c2)=O)O
M END