Mol:FL6DDAGI0001
From Metabolomics.JP
(Difference between revisions)
| Line 1: | Line 1: | ||
| − | + | ||
| − | + | ||
| − | Copyright: ARM project http://www.metabolome.jp/ | + | Copyright: ARM project http://www.metabolome.jp/ |
| − | 39 42 0 0 0 0 0 0 0 0999 V2000 | + | 39 42 0 0 0 0 0 0 0 0999 V2000 |
| − | -0.6213 -0.3475 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.6213 -0.3475 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.6213 -0.9899 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.6213 -0.9899 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.0650 -1.3111 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.0650 -1.3111 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.4913 -0.9899 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.4913 -0.9899 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.4913 -0.3475 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.4913 -0.3475 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.0650 -0.0263 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.0650 -0.0263 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.0476 -1.3111 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.0476 -1.3111 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.6039 -0.9899 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.6039 -0.9899 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.6039 -0.3475 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.6039 -0.3475 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.0476 -0.0263 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.0476 -0.0263 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.1600 -0.0264 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.1600 -0.0264 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.7270 -0.3538 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.7270 -0.3538 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.2939 -0.0264 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.2939 -0.0264 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.2939 0.6282 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.2939 0.6282 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.7270 0.9556 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.7270 0.9556 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.1600 0.6282 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.1600 0.6282 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.0678 -1.4538 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.0678 -1.4538 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.0650 0.6160 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.0650 0.6160 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.6213 0.9372 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.6213 0.9372 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.1776 -0.0263 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.1776 -0.0263 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.6213 1.5793 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.6213 1.5793 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.1774 1.9004 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.1774 1.9004 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.0652 1.9004 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.0652 1.9004 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.2344 -0.1855 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.2344 -0.1855 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.8632 -0.6755 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.8632 -0.6755 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.3287 -0.4676 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.3287 -0.4676 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.8130 -0.4620 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.8130 -0.4620 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.1877 -0.0872 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.1877 -0.0872 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.6554 -0.3340 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.6554 -0.3340 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.7482 -0.4822 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.7482 -0.4822 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.4419 -0.7036 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.4419 -0.7036 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.0225 -0.9817 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.0225 -0.9817 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.7482 0.8905 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.7482 0.8905 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.4732 -1.4879 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.4732 -1.4879 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.2413 -1.9004 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.2413 -1.9004 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.6112 -1.4716 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.6112 -1.4716 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.3256 -1.8841 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.3256 -1.8841 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.0538 0.4502 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.0538 0.4502 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.2569 0.2367 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.2569 0.2367 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1 2 1 0 0 0 0 | + | 1 2 1 0 0 0 0 |
| − | 2 3 2 0 0 0 0 | + | 2 3 2 0 0 0 0 |
| − | 3 4 1 0 0 0 0 | + | 3 4 1 0 0 0 0 |
| − | 4 5 2 0 0 0 0 | + | 4 5 2 0 0 0 0 |
| − | 5 6 1 0 0 0 0 | + | 5 6 1 0 0 0 0 |
| − | 6 1 2 0 0 0 0 | + | 6 1 2 0 0 0 0 |
| − | 4 7 1 0 0 0 0 | + | 4 7 1 0 0 0 0 |
| − | 7 8 1 0 0 0 0 | + | 7 8 1 0 0 0 0 |
| − | 8 9 1 0 0 0 0 | + | 8 9 1 0 0 0 0 |
| − | 9 10 1 0 0 0 0 | + | 9 10 1 0 0 0 0 |
| − | 10 5 1 0 0 0 0 | + | 10 5 1 0 0 0 0 |
| − | 9 11 1 6 0 0 0 | + | 9 11 1 6 0 0 0 |
| − | 11 12 2 0 0 0 0 | + | 11 12 2 0 0 0 0 |
| − | 12 13 1 0 0 0 0 | + | 12 13 1 0 0 0 0 |
| − | 13 14 2 0 0 0 0 | + | 13 14 2 0 0 0 0 |
| − | 14 15 1 0 0 0 0 | + | 14 15 1 0 0 0 0 |
| − | 15 16 2 0 0 0 0 | + | 15 16 2 0 0 0 0 |
| − | 16 11 1 0 0 0 0 | + | 16 11 1 0 0 0 0 |
| − | 8 17 1 1 0 0 0 | + | 8 17 1 1 0 0 0 |
| − | 6 18 1 0 0 0 0 | + | 6 18 1 0 0 0 0 |
| − | 18 19 1 0 0 0 0 | + | 18 19 1 0 0 0 0 |
| − | 20 1 1 0 0 0 0 | + | 20 1 1 0 0 0 0 |
| − | 19 21 2 0 0 0 0 | + | 19 21 2 0 0 0 0 |
| − | 21 22 1 0 0 0 0 | + | 21 22 1 0 0 0 0 |
| − | 21 23 1 0 0 0 0 | + | 21 23 1 0 0 0 0 |
| − | 24 25 1 1 0 0 0 | + | 24 25 1 1 0 0 0 |
| − | 25 26 1 1 0 0 0 | + | 25 26 1 1 0 0 0 |
| − | 27 26 1 1 0 0 0 | + | 27 26 1 1 0 0 0 |
| − | 27 28 1 0 0 0 0 | + | 27 28 1 0 0 0 0 |
| − | 28 29 1 0 0 0 0 | + | 28 29 1 0 0 0 0 |
| − | 29 24 1 0 0 0 0 | + | 29 24 1 0 0 0 0 |
| − | 24 30 1 0 0 0 0 | + | 24 30 1 0 0 0 0 |
| − | 25 31 1 0 0 0 0 | + | 25 31 1 0 0 0 0 |
| − | 26 32 1 0 0 0 0 | + | 26 32 1 0 0 0 0 |
| − | 27 20 1 0 0 0 0 | + | 27 20 1 0 0 0 0 |
| − | 14 33 1 0 0 0 0 | + | 14 33 1 0 0 0 0 |
| − | 3 34 1 0 0 0 0 | + | 3 34 1 0 0 0 0 |
| − | 34 35 1 0 0 0 0 | + | 34 35 1 0 0 0 0 |
| − | 7 36 1 1 0 0 0 | + | 7 36 1 1 0 0 0 |
| − | 36 37 1 0 0 0 0 | + | 36 37 1 0 0 0 0 |
| − | 29 38 1 0 0 0 0 | + | 29 38 1 0 0 0 0 |
| − | 38 39 1 0 0 0 0 | + | 38 39 1 0 0 0 0 |
| − | M STY 1 1 SUP | + | M STY 1 1 SUP |
| − | M SLB 1 1 1 | + | M SLB 1 1 1 |
| − | M SAL 1 2 34 35 | + | M SAL 1 2 34 35 |
| − | M SBL 1 1 37 | + | M SBL 1 1 37 |
| − | M SMT 1 OCH3 | + | M SMT 1 OCH3 |
| − | M SBV 1 37 -6.3466 3.3738 | + | M SBV 1 37 -6.3466 3.3738 |
| − | M STY 1 2 SUP | + | M STY 1 2 SUP |
| − | M SLB 1 2 2 | + | M SLB 1 2 2 |
| − | M SAL 2 2 36 37 | + | M SAL 2 2 36 37 |
| − | M SBL 2 1 39 | + | M SBL 2 1 39 |
| − | M SMT 2 OCH3 | + | M SMT 2 OCH3 |
| − | M SBV 2 39 -6.3749 3.3901 | + | M SBV 2 39 -6.3749 3.3901 |
| − | M STY 1 3 SUP | + | M STY 1 3 SUP |
| − | M SLB 1 3 3 | + | M SLB 1 3 3 |
| − | M SAL 3 2 38 39 | + | M SAL 3 2 38 39 |
| − | M SBL 3 1 41 | + | M SBL 3 1 41 |
| − | M SMT 3 ^CH2OH | + | M SMT 3 ^CH2OH |
| − | M SBV 3 41 -6.3369 4.3348 | + | M SBV 3 41 -6.3369 4.3348 |
| − | S SKP 8 | + | S SKP 8 |
| − | ID FL6DDAGI0001 | + | ID FL6DDAGI0001 |
| − | KNApSAcK_ID C00009032 | + | KNApSAcK_ID C00009032 |
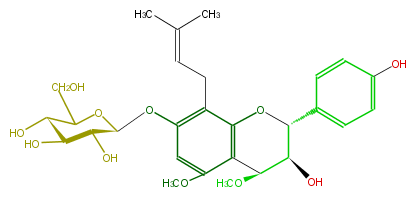
| − | NAME 3,7-Dihydroxy-4,5-dimethoxy-8-prenylflavan 7-O-beta-D-glucopyranoside | + | NAME 3,7-Dihydroxy-4,5-dimethoxy-8-prenylflavan 7-O-beta-D-glucopyranoside |
| − | CAS_RN 118555-86-5 | + | CAS_RN 118555-86-5 |
| − | FORMULA C28H36O11 | + | FORMULA C28H36O11 |
| − | EXACTMASS 548.225761994 | + | EXACTMASS 548.225761994 |
| − | AVERAGEMASS 548.57884 | + | AVERAGEMASS 548.57884 |
| − | SMILES C(Oc(c2)c(CC=C(C)C)c(O3)c(C(OC)C(O)C3c(c4)ccc(O)c4)c(OC)2)(O1)C(O)C(O)C(C(CO)1)O | + | SMILES C(Oc(c2)c(CC=C(C)C)c(O3)c(C(OC)C(O)C3c(c4)ccc(O)c4)c(OC)2)(O1)C(O)C(O)C(C(CO)1)O |
M END | M END | ||
| − | |||
Latest revision as of 09:00, 14 March 2009
Copyright: ARM project http://www.metabolome.jp/
39 42 0 0 0 0 0 0 0 0999 V2000
-0.6213 -0.3475 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.6213 -0.9899 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.0650 -1.3111 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.4913 -0.9899 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.4913 -0.3475 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.0650 -0.0263 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.0476 -1.3111 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.6039 -0.9899 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.6039 -0.3475 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.0476 -0.0263 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.1600 -0.0264 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.7270 -0.3538 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.2939 -0.0264 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.2939 0.6282 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.7270 0.9556 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.1600 0.6282 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.0678 -1.4538 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.0650 0.6160 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.6213 0.9372 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.1776 -0.0263 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.6213 1.5793 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.1774 1.9004 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.0652 1.9004 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.2344 -0.1855 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.8632 -0.6755 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.3287 -0.4676 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.8130 -0.4620 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.1877 -0.0872 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.6554 -0.3340 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.7482 -0.4822 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.4419 -0.7036 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.0225 -0.9817 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.7482 0.8905 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.4732 -1.4879 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.2413 -1.9004 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.6112 -1.4716 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.3256 -1.8841 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.0538 0.4502 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.2569 0.2367 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1 2 1 0 0 0 0
2 3 2 0 0 0 0
3 4 1 0 0 0 0
4 5 2 0 0 0 0
5 6 1 0 0 0 0
6 1 2 0 0 0 0
4 7 1 0 0 0 0
7 8 1 0 0 0 0
8 9 1 0 0 0 0
9 10 1 0 0 0 0
10 5 1 0 0 0 0
9 11 1 6 0 0 0
11 12 2 0 0 0 0
12 13 1 0 0 0 0
13 14 2 0 0 0 0
14 15 1 0 0 0 0
15 16 2 0 0 0 0
16 11 1 0 0 0 0
8 17 1 1 0 0 0
6 18 1 0 0 0 0
18 19 1 0 0 0 0
20 1 1 0 0 0 0
19 21 2 0 0 0 0
21 22 1 0 0 0 0
21 23 1 0 0 0 0
24 25 1 1 0 0 0
25 26 1 1 0 0 0
27 26 1 1 0 0 0
27 28 1 0 0 0 0
28 29 1 0 0 0 0
29 24 1 0 0 0 0
24 30 1 0 0 0 0
25 31 1 0 0 0 0
26 32 1 0 0 0 0
27 20 1 0 0 0 0
14 33 1 0 0 0 0
3 34 1 0 0 0 0
34 35 1 0 0 0 0
7 36 1 1 0 0 0
36 37 1 0 0 0 0
29 38 1 0 0 0 0
38 39 1 0 0 0 0
M STY 1 1 SUP
M SLB 1 1 1
M SAL 1 2 34 35
M SBL 1 1 37
M SMT 1 OCH3
M SBV 1 37 -6.3466 3.3738
M STY 1 2 SUP
M SLB 1 2 2
M SAL 2 2 36 37
M SBL 2 1 39
M SMT 2 OCH3
M SBV 2 39 -6.3749 3.3901
M STY 1 3 SUP
M SLB 1 3 3
M SAL 3 2 38 39
M SBL 3 1 41
M SMT 3 ^CH2OH
M SBV 3 41 -6.3369 4.3348
S SKP 8
ID FL6DDAGI0001
KNApSAcK_ID C00009032
NAME 3,7-Dihydroxy-4,5-dimethoxy-8-prenylflavan 7-O-beta-D-glucopyranoside
CAS_RN 118555-86-5
FORMULA C28H36O11
EXACTMASS 548.225761994
AVERAGEMASS 548.57884
SMILES C(Oc(c2)c(CC=C(C)C)c(O3)c(C(OC)C(O)C3c(c4)ccc(O)c4)c(OC)2)(O1)C(O)C(O)C(C(CO)1)O
M END