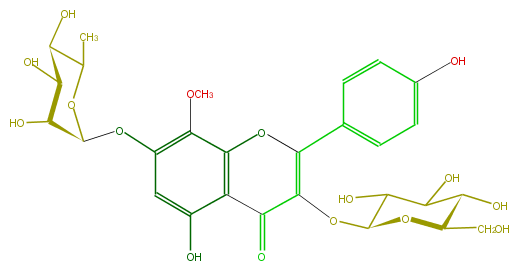
Mol:FL5FFAGL0009
From Metabolomics.JP
(Difference between revisions)
| Line 1: | Line 1: | ||
| − | + | ||
| − | + | ||
| − | Copyright: ARM project http://www.metabolome.jp/ | + | Copyright: ARM project http://www.metabolome.jp/ |
| − | 44 48 0 0 0 0 0 0 0 0999 V2000 | + | 44 48 0 0 0 0 0 0 0 0999 V2000 |
| − | -2.0774 -0.0071 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.0774 -0.0071 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.0774 -0.8322 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.0774 -0.8322 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.3628 -1.2446 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.3628 -1.2446 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.6483 -0.8322 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.6483 -0.8322 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.6483 -0.0071 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.6483 -0.0071 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.3628 0.4055 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.3628 0.4055 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.0662 -1.2446 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.0662 -1.2446 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.7807 -0.8322 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.7807 -0.8322 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.7807 -0.0071 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.7807 -0.0071 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.0662 0.4055 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.0662 0.4055 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.0662 -2.0661 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.0662 -2.0661 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.6804 0.5642 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.6804 0.5642 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.4086 0.1439 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.4086 0.1439 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.1368 0.5642 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.1368 0.5642 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.1368 1.4051 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.1368 1.4051 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.4086 1.8255 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.4086 1.8255 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.6804 1.4051 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.6804 1.4051 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.3628 -2.0694 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.3628 -2.0694 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.9162 1.8551 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.9162 1.8551 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.5042 -1.3581 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.5042 -1.3581 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.7855 0.4509 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.7855 0.4509 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.5748 -0.8484 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.5748 -0.8484 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.1885 -1.5176 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.1885 -1.5176 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.9315 -1.3053 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.9315 -1.3053 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.6790 -1.5176 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.6790 -1.5176 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.0654 -0.8484 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.0654 -0.8484 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.3224 -1.0607 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.3224 -1.0607 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.8174 -0.9183 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.8174 -0.9183 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.7895 -0.4938 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.7895 -0.4938 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.7612 -1.0517 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.7612 -1.0517 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.1927 0.6157 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.1927 0.6157 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.5181 0.2261 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.5181 0.2261 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.7321 0.9752 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.7321 0.9752 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.5181 1.7286 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.5181 1.7286 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.1927 2.1182 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.1927 2.1182 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.9787 1.3693 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.9787 1.3693 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.8770 2.7882 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.8770 2.7882 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.5083 2.3209 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.5083 2.3209 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.6253 1.8191 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.6253 1.8191 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.7612 0.6060 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.7612 0.6060 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.4488 -1.5040 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.4488 -1.5040 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.4216 -2.7882 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.4216 -2.7882 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.3628 1.1945 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.3628 1.1945 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.8022 2.3501 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.8022 2.3501 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1 2 1 0 0 0 0 | + | 1 2 1 0 0 0 0 |
| − | 2 3 2 0 0 0 0 | + | 2 3 2 0 0 0 0 |
| − | 4 5 2 0 0 0 0 | + | 4 5 2 0 0 0 0 |
| − | 5 6 1 0 0 0 0 | + | 5 6 1 0 0 0 0 |
| − | 6 1 2 0 0 0 0 | + | 6 1 2 0 0 0 0 |
| − | 4 7 1 0 0 0 0 | + | 4 7 1 0 0 0 0 |
| − | 7 8 1 0 0 0 0 | + | 7 8 1 0 0 0 0 |
| − | 8 9 2 0 0 0 0 | + | 8 9 2 0 0 0 0 |
| − | 9 10 1 0 0 0 0 | + | 9 10 1 0 0 0 0 |
| − | 10 5 1 0 0 0 0 | + | 10 5 1 0 0 0 0 |
| − | 7 11 2 0 0 0 0 | + | 7 11 2 0 0 0 0 |
| − | 9 12 1 0 0 0 0 | + | 9 12 1 0 0 0 0 |
| − | 12 13 2 0 0 0 0 | + | 12 13 2 0 0 0 0 |
| − | 13 14 1 0 0 0 0 | + | 13 14 1 0 0 0 0 |
| − | 14 15 2 0 0 0 0 | + | 14 15 2 0 0 0 0 |
| − | 15 16 1 0 0 0 0 | + | 15 16 1 0 0 0 0 |
| − | 16 17 2 0 0 0 0 | + | 16 17 2 0 0 0 0 |
| − | 17 12 1 0 0 0 0 | + | 17 12 1 0 0 0 0 |
| − | 3 18 1 0 0 0 0 | + | 3 18 1 0 0 0 0 |
| − | 19 15 1 0 0 0 0 | + | 19 15 1 0 0 0 0 |
| − | 20 8 1 0 0 0 0 | + | 20 8 1 0 0 0 0 |
| − | 4 3 1 0 0 0 0 | + | 4 3 1 0 0 0 0 |
| − | 1 21 1 0 0 0 0 | + | 1 21 1 0 0 0 0 |
| − | 22 23 1 0 0 0 0 | + | 22 23 1 0 0 0 0 |
| − | 23 24 1 1 0 0 0 | + | 23 24 1 1 0 0 0 |
| − | 24 25 1 1 0 0 0 | + | 24 25 1 1 0 0 0 |
| − | 26 25 1 1 0 0 0 | + | 26 25 1 1 0 0 0 |
| − | 26 27 1 0 0 0 0 | + | 26 27 1 0 0 0 0 |
| − | 27 22 1 0 0 0 0 | + | 27 22 1 0 0 0 0 |
| − | 22 28 1 0 0 0 0 | + | 22 28 1 0 0 0 0 |
| − | 27 29 1 0 0 0 0 | + | 27 29 1 0 0 0 0 |
| − | 26 30 1 0 0 0 0 | + | 26 30 1 0 0 0 0 |
| − | 23 20 1 0 0 0 0 | + | 23 20 1 0 0 0 0 |
| − | 32 31 1 1 0 0 0 | + | 32 31 1 1 0 0 0 |
| − | 32 33 1 0 0 0 0 | + | 32 33 1 0 0 0 0 |
| − | 33 34 1 0 0 0 0 | + | 33 34 1 0 0 0 0 |
| − | 34 35 1 0 0 0 0 | + | 34 35 1 0 0 0 0 |
| − | 35 36 1 1 0 0 0 | + | 35 36 1 1 0 0 0 |
| − | 36 31 1 1 0 0 0 | + | 36 31 1 1 0 0 0 |
| − | 35 37 1 0 0 0 0 | + | 35 37 1 0 0 0 0 |
| − | 34 38 1 0 0 0 0 | + | 34 38 1 0 0 0 0 |
| − | 36 39 1 0 0 0 0 | + | 36 39 1 0 0 0 0 |
| − | 31 40 1 0 0 0 0 | + | 31 40 1 0 0 0 0 |
| − | 32 21 1 0 0 0 0 | + | 32 21 1 0 0 0 0 |
| − | 41 42 1 0 0 0 0 | + | 41 42 1 0 0 0 0 |
| − | 25 41 1 0 0 0 0 | + | 25 41 1 0 0 0 0 |
| − | 43 44 1 0 0 0 0 | + | 43 44 1 0 0 0 0 |
| − | 6 43 1 0 0 0 0 | + | 6 43 1 0 0 0 0 |
| − | M STY 1 1 SUP | + | M STY 1 1 SUP |
| − | M SLB 1 1 1 | + | M SLB 1 1 1 |
| − | M SAL 1 2 41 42 | + | M SAL 1 2 41 42 |
| − | M SBL 1 1 46 | + | M SBL 1 1 46 |
| − | M SMT 1 CH2OH | + | M SMT 1 CH2OH |
| − | M SBV 1 46 -0.7698 -0.0135 | + | M SBV 1 46 -0.7698 -0.0135 |
| − | M STY 1 2 SUP | + | M STY 1 2 SUP |
| − | M SLB 1 2 2 | + | M SLB 1 2 2 |
| − | M SAL 2 2 43 44 | + | M SAL 2 2 43 44 |
| − | M SBL 2 1 48 | + | M SBL 2 1 48 |
| − | M SMT 2 OCH3 | + | M SMT 2 OCH3 |
| − | M SBV 2 48 0.0000 -0.7890 | + | M SBV 2 48 0.0000 -0.7890 |
| − | S SKP 5 | + | S SKP 5 |
| − | ID FL5FFAGL0009 | + | ID FL5FFAGL0009 |
| − | FORMULA C28H32O16 | + | FORMULA C28H32O16 |
| − | EXACTMASS 624.1690349759999 | + | EXACTMASS 624.1690349759999 |
| − | AVERAGEMASS 624.54408 | + | AVERAGEMASS 624.54408 |
| − | SMILES OC(C5CO)C(O)C(O)C(O5)OC(=C3c(c4)ccc(c4)O)C(c(c2O)c(O3)c(OC)c(c2)OC(C1O)OC(C(O)C1O)C)=O | + | SMILES OC(C5CO)C(O)C(O)C(O5)OC(=C3c(c4)ccc(c4)O)C(c(c2O)c(O3)c(OC)c(c2)OC(C1O)OC(C(O)C1O)C)=O |
M END | M END | ||
| − | |||
Latest revision as of 09:00, 14 March 2009
Copyright: ARM project http://www.metabolome.jp/
44 48 0 0 0 0 0 0 0 0999 V2000
-2.0774 -0.0071 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.0774 -0.8322 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.3628 -1.2446 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.6483 -0.8322 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.6483 -0.0071 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.3628 0.4055 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.0662 -1.2446 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.7807 -0.8322 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.7807 -0.0071 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.0662 0.4055 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.0662 -2.0661 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.6804 0.5642 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.4086 0.1439 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.1368 0.5642 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.1368 1.4051 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.4086 1.8255 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.6804 1.4051 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.3628 -2.0694 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.9162 1.8551 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.5042 -1.3581 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.7855 0.4509 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.5748 -0.8484 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.1885 -1.5176 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.9315 -1.3053 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.6790 -1.5176 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.0654 -0.8484 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.3224 -1.0607 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.8174 -0.9183 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.7895 -0.4938 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
4.7612 -1.0517 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.1927 0.6157 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.5181 0.2261 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.7321 0.9752 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.5181 1.7286 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.1927 2.1182 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.9787 1.3693 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.8770 2.7882 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.5083 2.3209 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.6253 1.8191 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.7612 0.6060 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
4.4488 -1.5040 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.4216 -2.7882 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.3628 1.1945 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.8022 2.3501 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1 2 1 0 0 0 0
2 3 2 0 0 0 0
4 5 2 0 0 0 0
5 6 1 0 0 0 0
6 1 2 0 0 0 0
4 7 1 0 0 0 0
7 8 1 0 0 0 0
8 9 2 0 0 0 0
9 10 1 0 0 0 0
10 5 1 0 0 0 0
7 11 2 0 0 0 0
9 12 1 0 0 0 0
12 13 2 0 0 0 0
13 14 1 0 0 0 0
14 15 2 0 0 0 0
15 16 1 0 0 0 0
16 17 2 0 0 0 0
17 12 1 0 0 0 0
3 18 1 0 0 0 0
19 15 1 0 0 0 0
20 8 1 0 0 0 0
4 3 1 0 0 0 0
1 21 1 0 0 0 0
22 23 1 0 0 0 0
23 24 1 1 0 0 0
24 25 1 1 0 0 0
26 25 1 1 0 0 0
26 27 1 0 0 0 0
27 22 1 0 0 0 0
22 28 1 0 0 0 0
27 29 1 0 0 0 0
26 30 1 0 0 0 0
23 20 1 0 0 0 0
32 31 1 1 0 0 0
32 33 1 0 0 0 0
33 34 1 0 0 0 0
34 35 1 0 0 0 0
35 36 1 1 0 0 0
36 31 1 1 0 0 0
35 37 1 0 0 0 0
34 38 1 0 0 0 0
36 39 1 0 0 0 0
31 40 1 0 0 0 0
32 21 1 0 0 0 0
41 42 1 0 0 0 0
25 41 1 0 0 0 0
43 44 1 0 0 0 0
6 43 1 0 0 0 0
M STY 1 1 SUP
M SLB 1 1 1
M SAL 1 2 41 42
M SBL 1 1 46
M SMT 1 CH2OH
M SBV 1 46 -0.7698 -0.0135
M STY 1 2 SUP
M SLB 1 2 2
M SAL 2 2 43 44
M SBL 2 1 48
M SMT 2 OCH3
M SBV 2 48 0.0000 -0.7890
S SKP 5
ID FL5FFAGL0009
FORMULA C28H32O16
EXACTMASS 624.1690349759999
AVERAGEMASS 624.54408
SMILES OC(C5CO)C(O)C(O)C(O5)OC(=C3c(c4)ccc(c4)O)C(c(c2O)c(O3)c(OC)c(c2)OC(C1O)OC(C(O)C1O)C)=O
M END