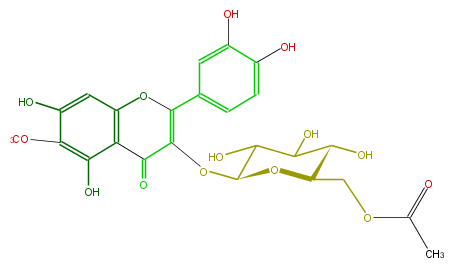
Mol:FL5FECGL0014
From Metabolomics.JP
(Difference between revisions)
| Line 1: | Line 1: | ||
| − | + | ||
| − | + | ||
| − | Copyright: ARM project http://www.metabolome.jp/ | + | Copyright: ARM project http://www.metabolome.jp/ |
| − | 38 41 0 0 0 0 0 0 0 0999 V2000 | + | 38 41 0 0 0 0 0 0 0 0999 V2000 |
| − | -3.3346 0.4406 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.3346 0.4406 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.3346 -0.2017 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.3346 -0.2017 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.7783 -0.5229 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.7783 -0.5229 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.2220 -0.2017 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.2220 -0.2017 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.2220 0.4406 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.2220 0.4406 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.7783 0.7618 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.7783 0.7618 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.6657 -0.5229 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.6657 -0.5229 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.1094 -0.2017 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.1094 -0.2017 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.1094 0.4406 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.1094 0.4406 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.6657 0.7618 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.6657 0.7618 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.6657 -1.0238 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.6657 -1.0238 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.5533 0.7617 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.5533 0.7617 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.0136 0.4343 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.0136 0.4343 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.5806 0.7617 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.5806 0.7617 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.5806 1.4164 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.5806 1.4164 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.0136 1.7437 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.0136 1.7437 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.5533 1.4164 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.5533 1.4164 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.4613 -0.7520 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.4613 -0.7520 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.5841 -0.2606 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.5841 -0.2606 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.1965 -0.9320 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.1965 -0.9320 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.9419 -0.7190 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.9419 -0.7190 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.6919 -0.9320 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.6919 -0.9320 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.0796 -0.2606 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.0796 -0.2606 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.3341 -0.4736 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.3341 -0.4736 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.1430 -0.4554 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.1430 -0.4554 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.6029 -0.0080 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.6029 -0.0080 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.6939 -0.4252 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.6939 -0.4252 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.1474 1.7436 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.1474 1.7436 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.7783 -1.1650 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.7783 -1.1650 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.9456 0.6391 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.9456 0.6391 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.3208 -0.9320 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.3208 -0.9320 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.8116 -1.6837 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.8116 -1.6837 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.6366 -1.6837 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.6366 -1.6837 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.0264 -1.0085 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.0264 -1.0085 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.0136 2.3982 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.0136 2.3982 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.0491 -2.3982 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.0491 -2.3982 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.0491 -0.1103 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.0491 -0.1103 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.3346 -0.5228 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.3346 -0.5228 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1 2 1 0 0 0 0 | + | 1 2 1 0 0 0 0 |
| − | 2 3 2 0 0 0 0 | + | 2 3 2 0 0 0 0 |
| − | 3 4 1 0 0 0 0 | + | 3 4 1 0 0 0 0 |
| − | 4 5 2 0 0 0 0 | + | 4 5 2 0 0 0 0 |
| − | 5 6 1 0 0 0 0 | + | 5 6 1 0 0 0 0 |
| − | 6 1 2 0 0 0 0 | + | 6 1 2 0 0 0 0 |
| − | 4 7 1 0 0 0 0 | + | 4 7 1 0 0 0 0 |
| − | 7 8 1 0 0 0 0 | + | 7 8 1 0 0 0 0 |
| − | 8 9 2 0 0 0 0 | + | 8 9 2 0 0 0 0 |
| − | 9 10 1 0 0 0 0 | + | 9 10 1 0 0 0 0 |
| − | 10 5 1 0 0 0 0 | + | 10 5 1 0 0 0 0 |
| − | 7 11 2 0 0 0 0 | + | 7 11 2 0 0 0 0 |
| − | 9 12 1 0 0 0 0 | + | 9 12 1 0 0 0 0 |
| − | 12 13 2 0 0 0 0 | + | 12 13 2 0 0 0 0 |
| − | 13 14 1 0 0 0 0 | + | 13 14 1 0 0 0 0 |
| − | 14 15 2 0 0 0 0 | + | 14 15 2 0 0 0 0 |
| − | 15 16 1 0 0 0 0 | + | 15 16 1 0 0 0 0 |
| − | 16 17 2 0 0 0 0 | + | 16 17 2 0 0 0 0 |
| − | 17 12 1 0 0 0 0 | + | 17 12 1 0 0 0 0 |
| − | 19 20 1 0 0 0 0 | + | 19 20 1 0 0 0 0 |
| − | 20 21 1 1 0 0 0 | + | 20 21 1 1 0 0 0 |
| − | 23 22 1 1 0 0 0 | + | 23 22 1 1 0 0 0 |
| − | 23 24 1 0 0 0 0 | + | 23 24 1 0 0 0 0 |
| − | 24 19 1 0 0 0 0 | + | 24 19 1 0 0 0 0 |
| − | 19 25 1 0 0 0 0 | + | 19 25 1 0 0 0 0 |
| − | 24 26 1 0 0 0 0 | + | 24 26 1 0 0 0 0 |
| − | 23 27 1 0 0 0 0 | + | 23 27 1 0 0 0 0 |
| − | 20 18 1 0 0 0 0 | + | 20 18 1 0 0 0 0 |
| − | 18 8 1 0 0 0 0 | + | 18 8 1 0 0 0 0 |
| − | 22 21 1 1 0 0 0 | + | 22 21 1 1 0 0 0 |
| − | 15 28 1 0 0 0 0 | + | 15 28 1 0 0 0 0 |
| − | 3 29 1 0 0 0 0 | + | 3 29 1 0 0 0 0 |
| − | 1 30 1 0 0 0 0 | + | 1 30 1 0 0 0 0 |
| − | 22 31 1 0 0 0 0 | + | 22 31 1 0 0 0 0 |
| − | 31 32 1 0 0 0 0 | + | 31 32 1 0 0 0 0 |
| − | 32 33 1 0 0 0 0 | + | 32 33 1 0 0 0 0 |
| − | 33 34 2 0 0 0 0 | + | 33 34 2 0 0 0 0 |
| − | 16 35 1 0 0 0 0 | + | 16 35 1 0 0 0 0 |
| − | 33 36 1 0 0 0 0 | + | 33 36 1 0 0 0 0 |
| − | 2 37 1 0 0 0 0 | + | 2 37 1 0 0 0 0 |
| − | 37 38 1 0 0 0 0 | + | 37 38 1 0 0 0 0 |
| − | M STY 1 1 SUP | + | M STY 1 1 SUP |
| − | M SLB 1 1 1 | + | M SLB 1 1 1 |
| − | M SAL 1 2 37 38 | + | M SAL 1 2 37 38 |
| − | M SBL 1 1 40 | + | M SBL 1 1 40 |
| − | M SMT 1 ^OCH3 | + | M SMT 1 ^OCH3 |
| − | M SBV 1 40 -6.4581 5.9977 | + | M SBV 1 40 -6.4581 5.9977 |
| − | S SKP 8 | + | S SKP 8 |
| − | ID FL5FECGL0014 | + | ID FL5FECGL0014 |
| − | KNApSAcK_ID C00006021 | + | KNApSAcK_ID C00006021 |
| − | NAME Patuletin 3-(6''-acetylglucoside) | + | NAME Patuletin 3-(6''-acetylglucoside) |
| − | CAS_RN 118201-24-4 | + | CAS_RN 118201-24-4 |
| − | FORMULA C24H24O14 | + | FORMULA C24H24O14 |
| − | EXACTMASS 536.116605476 | + | EXACTMASS 536.116605476 |
| − | AVERAGEMASS 536.43896 | + | AVERAGEMASS 536.43896 |
| − | SMILES OC(C4COC(C)=O)C(O)C(O)C(O4)OC(C(=O)1)=C(c(c3)cc(c(c3)O)O)Oc(c2)c(c(c(OC)c(O)2)O)1 | + | SMILES OC(C4COC(C)=O)C(O)C(O)C(O4)OC(C(=O)1)=C(c(c3)cc(c(c3)O)O)Oc(c2)c(c(c(OC)c(O)2)O)1 |
M END | M END | ||
| − | |||
Latest revision as of 09:00, 14 March 2009
Copyright: ARM project http://www.metabolome.jp/
38 41 0 0 0 0 0 0 0 0999 V2000
-3.3346 0.4406 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.3346 -0.2017 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.7783 -0.5229 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.2220 -0.2017 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.2220 0.4406 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.7783 0.7618 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.6657 -0.5229 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.1094 -0.2017 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.1094 0.4406 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.6657 0.7618 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.6657 -1.0238 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.5533 0.7617 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.0136 0.4343 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.5806 0.7617 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.5806 1.4164 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.0136 1.7437 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.5533 1.4164 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.4613 -0.7520 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.5841 -0.2606 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.1965 -0.9320 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.9419 -0.7190 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.6919 -0.9320 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.0796 -0.2606 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.3341 -0.4736 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.1430 -0.4554 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.6029 -0.0080 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.6939 -0.4252 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.1474 1.7436 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.7783 -1.1650 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.9456 0.6391 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.3208 -0.9320 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.8116 -1.6837 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.6366 -1.6837 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.0264 -1.0085 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.0136 2.3982 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
4.0491 -2.3982 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.0491 -0.1103 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.3346 -0.5228 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1 2 1 0 0 0 0
2 3 2 0 0 0 0
3 4 1 0 0 0 0
4 5 2 0 0 0 0
5 6 1 0 0 0 0
6 1 2 0 0 0 0
4 7 1 0 0 0 0
7 8 1 0 0 0 0
8 9 2 0 0 0 0
9 10 1 0 0 0 0
10 5 1 0 0 0 0
7 11 2 0 0 0 0
9 12 1 0 0 0 0
12 13 2 0 0 0 0
13 14 1 0 0 0 0
14 15 2 0 0 0 0
15 16 1 0 0 0 0
16 17 2 0 0 0 0
17 12 1 0 0 0 0
19 20 1 0 0 0 0
20 21 1 1 0 0 0
23 22 1 1 0 0 0
23 24 1 0 0 0 0
24 19 1 0 0 0 0
19 25 1 0 0 0 0
24 26 1 0 0 0 0
23 27 1 0 0 0 0
20 18 1 0 0 0 0
18 8 1 0 0 0 0
22 21 1 1 0 0 0
15 28 1 0 0 0 0
3 29 1 0 0 0 0
1 30 1 0 0 0 0
22 31 1 0 0 0 0
31 32 1 0 0 0 0
32 33 1 0 0 0 0
33 34 2 0 0 0 0
16 35 1 0 0 0 0
33 36 1 0 0 0 0
2 37 1 0 0 0 0
37 38 1 0 0 0 0
M STY 1 1 SUP
M SLB 1 1 1
M SAL 1 2 37 38
M SBL 1 1 40
M SMT 1 ^OCH3
M SBV 1 40 -6.4581 5.9977
S SKP 8
ID FL5FECGL0014
KNApSAcK_ID C00006021
NAME Patuletin 3-(6''-acetylglucoside)
CAS_RN 118201-24-4
FORMULA C24H24O14
EXACTMASS 536.116605476
AVERAGEMASS 536.43896
SMILES OC(C4COC(C)=O)C(O)C(O)C(O4)OC(C(=O)1)=C(c(c3)cc(c(c3)O)O)Oc(c2)c(c(c(OC)c(O)2)O)1
M END