Mol:FL5FAIGL0005
From Metabolomics.JP
(Difference between revisions)
| Line 1: | Line 1: | ||
| − | + | ||
| − | + | ||
| − | Copyright: ARM project http://www.metabolome.jp/ | + | Copyright: ARM project http://www.metabolome.jp/ |
| − | 42 45 0 0 0 0 0 0 0 0999 V2000 | + | 42 45 0 0 0 0 0 0 0 0999 V2000 |
| − | -2.1643 -0.7787 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.1643 -0.7787 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.4977 -0.3939 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.4977 -0.3939 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.4977 0.3758 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.4977 0.3758 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.1643 0.7607 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.1643 0.7607 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.8308 0.3758 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.8308 0.3758 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.8308 -0.3939 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.8308 -0.3939 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.8311 -0.7787 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.8311 -0.7787 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.1646 -0.3939 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.1646 -0.3939 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.1646 0.3758 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.1646 0.3758 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.8311 0.7607 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.8311 0.7607 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.4661 0.7400 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.4661 0.7400 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.1135 0.3662 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.1135 0.3662 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.7609 0.7400 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.7609 0.7400 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.7609 1.4875 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.7609 1.4875 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.1135 1.8612 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.1135 1.8612 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.4661 1.4875 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.4661 1.4875 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.3233 1.8122 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.3233 1.8122 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.1135 2.4596 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.1135 2.4596 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.5360 2.8821 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.5360 2.8821 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.4094 0.7099 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.4094 0.7099 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.1643 -1.3242 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.1643 -1.3242 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.8311 -1.3080 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.8311 -1.3080 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.6019 -0.8364 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.6019 -0.8364 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.6758 -1.0075 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.6758 -1.0075 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.8956 -1.2756 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.8956 -1.2756 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.6428 -1.6254 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.6428 -1.6254 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.1074 -2.3072 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.1074 -2.3072 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.8877 -2.0392 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.8877 -2.0392 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.1405 -1.6893 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.1405 -1.6893 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.7408 -2.4666 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.7408 -2.4666 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.8590 -2.8821 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.8590 -2.8821 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.1881 -2.5418 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.1881 -2.5418 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.5594 -1.4273 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.5594 -1.4273 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.9762 -1.3556 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.9762 -1.3556 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.1893 -0.8106 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.1893 -0.8106 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.4094 -1.6746 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.4094 -1.6746 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.3033 -0.7999 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.3033 -0.7999 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.3033 -0.2507 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.3033 -0.2507 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.8893 -0.0117 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.8893 -0.0117 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.6862 -0.0297 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.6862 -0.0297 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.3067 0.4248 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.3067 0.4248 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.9674 0.4248 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.9674 0.4248 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1 2 2 0 0 0 0 | + | 1 2 2 0 0 0 0 |
| − | 2 3 1 0 0 0 0 | + | 2 3 1 0 0 0 0 |
| − | 3 4 2 0 0 0 0 | + | 3 4 2 0 0 0 0 |
| − | 4 5 1 0 0 0 0 | + | 4 5 1 0 0 0 0 |
| − | 5 6 2 0 0 0 0 | + | 5 6 2 0 0 0 0 |
| − | 6 1 1 0 0 0 0 | + | 6 1 1 0 0 0 0 |
| − | 2 7 1 0 0 0 0 | + | 2 7 1 0 0 0 0 |
| − | 7 8 1 0 0 0 0 | + | 7 8 1 0 0 0 0 |
| − | 8 9 2 0 0 0 0 | + | 8 9 2 0 0 0 0 |
| − | 9 10 1 0 0 0 0 | + | 9 10 1 0 0 0 0 |
| − | 10 3 1 0 0 0 0 | + | 10 3 1 0 0 0 0 |
| − | 9 11 1 0 0 0 0 | + | 9 11 1 0 0 0 0 |
| − | 11 12 2 0 0 0 0 | + | 11 12 2 0 0 0 0 |
| − | 12 13 1 0 0 0 0 | + | 12 13 1 0 0 0 0 |
| − | 13 14 2 0 0 0 0 | + | 13 14 2 0 0 0 0 |
| − | 14 15 1 0 0 0 0 | + | 14 15 1 0 0 0 0 |
| − | 15 16 2 0 0 0 0 | + | 15 16 2 0 0 0 0 |
| − | 16 11 1 0 0 0 0 | + | 16 11 1 0 0 0 0 |
| − | 14 17 1 0 0 0 0 | + | 14 17 1 0 0 0 0 |
| − | 15 18 1 0 0 0 0 | + | 15 18 1 0 0 0 0 |
| − | 18 19 1 0 0 0 0 | + | 18 19 1 0 0 0 0 |
| − | 5 20 1 0 0 0 0 | + | 5 20 1 0 0 0 0 |
| − | 1 21 1 0 0 0 0 | + | 1 21 1 0 0 0 0 |
| − | 7 22 2 0 0 0 0 | + | 7 22 2 0 0 0 0 |
| − | 8 23 1 0 0 0 0 | + | 8 23 1 0 0 0 0 |
| − | 24 25 1 0 0 0 0 | + | 24 25 1 0 0 0 0 |
| − | 25 26 1 1 0 0 0 | + | 25 26 1 1 0 0 0 |
| − | 27 26 1 1 0 0 0 | + | 27 26 1 1 0 0 0 |
| − | 28 27 1 1 0 0 0 | + | 28 27 1 1 0 0 0 |
| − | 28 29 1 0 0 0 0 | + | 28 29 1 0 0 0 0 |
| − | 29 24 1 0 0 0 0 | + | 29 24 1 0 0 0 0 |
| − | 23 25 1 0 0 0 0 | + | 23 25 1 0 0 0 0 |
| − | 27 30 1 0 0 0 0 | + | 27 30 1 0 0 0 0 |
| − | 30 31 1 0 0 0 0 | + | 30 31 1 0 0 0 0 |
| − | 28 32 1 0 0 0 0 | + | 28 32 1 0 0 0 0 |
| − | 29 33 1 0 0 0 0 | + | 29 33 1 0 0 0 0 |
| − | 33 34 1 0 0 0 0 | + | 33 34 1 0 0 0 0 |
| − | 34 35 2 0 0 0 0 | + | 34 35 2 0 0 0 0 |
| − | 34 36 1 0 0 0 0 | + | 34 36 1 0 0 0 0 |
| − | 24 37 1 0 0 0 0 | + | 24 37 1 0 0 0 0 |
| − | 37 38 1 0 0 0 0 | + | 37 38 1 0 0 0 0 |
| − | 38 39 2 0 0 0 0 | + | 38 39 2 0 0 0 0 |
| − | 38 40 1 0 0 0 0 | + | 38 40 1 0 0 0 0 |
| − | 13 41 1 0 0 0 0 | + | 13 41 1 0 0 0 0 |
| − | 41 42 1 0 0 0 0 | + | 41 42 1 0 0 0 0 |
| − | S SKP 8 | + | S SKP 8 |
| − | ID FL5FAIGL0005 | + | ID FL5FAIGL0005 |
| − | KNApSAcK_ID C00011134 | + | KNApSAcK_ID C00011134 |
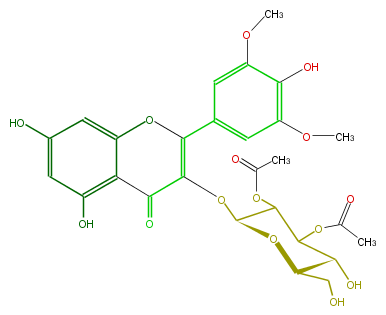
| − | NAME 3',5'-O-Dimethylmyricetin 3-O-beta-D-2'',3''-diacetylglucopyranoside;Syringetin 3-(2'',3''-diacetylglucoside);3-[(2,3-Di-O-acetyl-beta-D-glucopyranosyl)oxy]-5,7-dihydroxy-2-(4-hydroxy-3,5-dimethoxyphenyl)-4H-1-benzopyran-4-one | + | NAME 3',5'-O-Dimethylmyricetin 3-O-beta-D-2'',3''-diacetylglucopyranoside;Syringetin 3-(2'',3''-diacetylglucoside);3-[(2,3-Di-O-acetyl-beta-D-glucopyranosyl)oxy]-5,7-dihydroxy-2-(4-hydroxy-3,5-dimethoxyphenyl)-4H-1-benzopyran-4-one |
| − | CAS_RN 600736-89-8 | + | CAS_RN 600736-89-8 |
| − | FORMULA C27H28O15 | + | FORMULA C27H28O15 |
| − | EXACTMASS 592.1428202259999 | + | EXACTMASS 592.1428202259999 |
| − | AVERAGEMASS 592.5022200000001 | + | AVERAGEMASS 592.5022200000001 |
| − | SMILES O(C(=C(c(c4)cc(c(O)c4OC)OC)2)C(=O)c(c3O)c(cc(c3)O)O2)C(C1OC(C)=O)OC(C(C1OC(C)=O)O)CO | + | SMILES O(C(=C(c(c4)cc(c(O)c4OC)OC)2)C(=O)c(c3O)c(cc(c3)O)O2)C(C1OC(C)=O)OC(C(C1OC(C)=O)O)CO |
M END | M END | ||
| − | |||
Latest revision as of 09:00, 14 March 2009
Copyright: ARM project http://www.metabolome.jp/
42 45 0 0 0 0 0 0 0 0999 V2000
-2.1643 -0.7787 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.4977 -0.3939 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.4977 0.3758 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.1643 0.7607 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.8308 0.3758 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.8308 -0.3939 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.8311 -0.7787 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.1646 -0.3939 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.1646 0.3758 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.8311 0.7607 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.4661 0.7400 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.1135 0.3662 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.7609 0.7400 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.7609 1.4875 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.1135 1.8612 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.4661 1.4875 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.3233 1.8122 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.1135 2.4596 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.5360 2.8821 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.4094 0.7099 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.1643 -1.3242 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.8311 -1.3080 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.6019 -0.8364 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.6758 -1.0075 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.8956 -1.2756 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.6428 -1.6254 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.1074 -2.3072 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.8877 -2.0392 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.1405 -1.6893 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.7408 -2.4666 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.8590 -2.8821 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.1881 -2.5418 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.5594 -1.4273 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.9762 -1.3556 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.1893 -0.8106 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.4094 -1.6746 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.3033 -0.7999 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.3033 -0.2507 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.8893 -0.0117 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.6862 -0.0297 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.3067 0.4248 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.9674 0.4248 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1 2 2 0 0 0 0
2 3 1 0 0 0 0
3 4 2 0 0 0 0
4 5 1 0 0 0 0
5 6 2 0 0 0 0
6 1 1 0 0 0 0
2 7 1 0 0 0 0
7 8 1 0 0 0 0
8 9 2 0 0 0 0
9 10 1 0 0 0 0
10 3 1 0 0 0 0
9 11 1 0 0 0 0
11 12 2 0 0 0 0
12 13 1 0 0 0 0
13 14 2 0 0 0 0
14 15 1 0 0 0 0
15 16 2 0 0 0 0
16 11 1 0 0 0 0
14 17 1 0 0 0 0
15 18 1 0 0 0 0
18 19 1 0 0 0 0
5 20 1 0 0 0 0
1 21 1 0 0 0 0
7 22 2 0 0 0 0
8 23 1 0 0 0 0
24 25 1 0 0 0 0
25 26 1 1 0 0 0
27 26 1 1 0 0 0
28 27 1 1 0 0 0
28 29 1 0 0 0 0
29 24 1 0 0 0 0
23 25 1 0 0 0 0
27 30 1 0 0 0 0
30 31 1 0 0 0 0
28 32 1 0 0 0 0
29 33 1 0 0 0 0
33 34 1 0 0 0 0
34 35 2 0 0 0 0
34 36 1 0 0 0 0
24 37 1 0 0 0 0
37 38 1 0 0 0 0
38 39 2 0 0 0 0
38 40 1 0 0 0 0
13 41 1 0 0 0 0
41 42 1 0 0 0 0
S SKP 8
ID FL5FAIGL0005
KNApSAcK_ID C00011134
NAME 3',5'-O-Dimethylmyricetin 3-O-beta-D-2'',3''-diacetylglucopyranoside;Syringetin 3-(2'',3''-diacetylglucoside);3-[(2,3-Di-O-acetyl-beta-D-glucopyranosyl)oxy]-5,7-dihydroxy-2-(4-hydroxy-3,5-dimethoxyphenyl)-4H-1-benzopyran-4-one
CAS_RN 600736-89-8
FORMULA C27H28O15
EXACTMASS 592.1428202259999
AVERAGEMASS 592.5022200000001
SMILES O(C(=C(c(c4)cc(c(O)c4OC)OC)2)C(=O)c(c3O)c(cc(c3)O)O2)C(C1OC(C)=O)OC(C(C1OC(C)=O)O)CO
M END