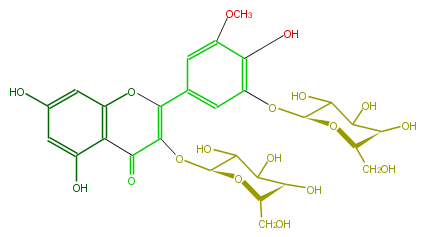
Mol:FL5FAHGL0003
From Metabolomics.JP
(Difference between revisions)
| Line 1: | Line 1: | ||
| − | + | ||
| − | + | ||
| − | Copyright: ARM project http://www.metabolome.jp/ | + | Copyright: ARM project http://www.metabolome.jp/ |
| − | 46 50 0 0 0 0 0 0 0 0999 V2000 | + | 46 50 0 0 0 0 0 0 0 0999 V2000 |
| − | -3.3587 0.2950 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.3587 0.2950 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.3587 -0.3474 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.3587 -0.3474 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.8024 -0.6686 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.8024 -0.6686 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.2461 -0.3474 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.2461 -0.3474 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.2461 0.2950 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.2461 0.2950 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.8024 0.6161 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.8024 0.6161 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.6898 -0.6686 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.6898 -0.6686 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.1335 -0.3474 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.1335 -0.3474 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.1335 0.2950 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.1335 0.2950 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.6898 0.6161 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.6898 0.6161 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.6898 -1.1694 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.6898 -1.1694 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.5774 0.6160 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.5774 0.6160 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.0105 0.2887 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.0105 0.2887 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.5565 0.6160 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.5565 0.6160 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.5565 1.2707 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.5565 1.2707 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.0105 1.5981 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.0105 1.5981 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.5774 1.2707 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.5774 1.2707 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.9148 0.6160 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.9148 0.6160 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.4225 1.7707 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.4225 1.7707 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.7394 -0.7363 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.7394 -0.7363 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.8024 -1.3107 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.8024 -1.3107 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.1233 0.2888 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.1233 0.2888 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.2861 0.3775 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 | + | 2.2861 0.3775 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 |
| − | 1.7650 0.0767 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | 1.7650 0.0767 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | 2.3487 -0.0694 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.3487 -0.0694 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.7700 -0.5036 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | 2.7700 -0.5036 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | 3.2911 -0.2028 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | 3.2911 -0.2028 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | 2.7074 -0.0567 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 | + | 2.7074 -0.0567 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 |
| − | 1.7274 0.5272 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.7274 0.5272 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.9807 0.3336 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.9807 0.3336 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.7752 -0.0731 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.7752 -0.0731 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.3886 -0.6744 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 | + | 0.3886 -0.6744 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 |
| − | -0.1325 -0.9752 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | -0.1325 -0.9752 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | 0.4512 -1.1213 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.4512 -1.1213 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.8725 -1.5554 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | 0.8725 -1.5554 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | 1.3936 -1.2547 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | 1.3936 -1.2547 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | 0.8099 -1.1085 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 | + | 0.8099 -1.1085 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 |
| − | -0.1701 -0.5247 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.1701 -0.5247 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.0832 -0.7182 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.0832 -0.7182 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.8871 -1.3417 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.8871 -1.3417 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.9090 -2.0313 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.9090 -2.0313 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.9090 -2.8562 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.9090 -2.8562 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.0043 -0.9194 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.0043 -0.9194 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.3529 -1.6670 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.3529 -1.6670 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.2729 2.1742 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.2729 2.1742 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.7143 3.0715 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.7143 3.0715 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1 2 1 0 0 0 0 | + | 1 2 1 0 0 0 0 |
| − | 2 3 2 0 0 0 0 | + | 2 3 2 0 0 0 0 |
| − | 3 4 1 0 0 0 0 | + | 3 4 1 0 0 0 0 |
| − | 4 5 2 0 0 0 0 | + | 4 5 2 0 0 0 0 |
| − | 5 6 1 0 0 0 0 | + | 5 6 1 0 0 0 0 |
| − | 6 1 2 0 0 0 0 | + | 6 1 2 0 0 0 0 |
| − | 4 7 1 0 0 0 0 | + | 4 7 1 0 0 0 0 |
| − | 7 8 1 0 0 0 0 | + | 7 8 1 0 0 0 0 |
| − | 8 9 2 0 0 0 0 | + | 8 9 2 0 0 0 0 |
| − | 9 10 1 0 0 0 0 | + | 9 10 1 0 0 0 0 |
| − | 10 5 1 0 0 0 0 | + | 10 5 1 0 0 0 0 |
| − | 7 11 2 0 0 0 0 | + | 7 11 2 0 0 0 0 |
| − | 9 12 1 0 0 0 0 | + | 9 12 1 0 0 0 0 |
| − | 12 13 2 0 0 0 0 | + | 12 13 2 0 0 0 0 |
| − | 13 14 1 0 0 0 0 | + | 13 14 1 0 0 0 0 |
| − | 14 15 2 0 0 0 0 | + | 14 15 2 0 0 0 0 |
| − | 15 16 1 0 0 0 0 | + | 15 16 1 0 0 0 0 |
| − | 16 17 2 0 0 0 0 | + | 16 17 2 0 0 0 0 |
| − | 17 12 1 0 0 0 0 | + | 17 12 1 0 0 0 0 |
| − | 1 18 1 0 0 0 0 | + | 1 18 1 0 0 0 0 |
| − | 15 19 1 0 0 0 0 | + | 15 19 1 0 0 0 0 |
| − | 8 20 1 0 0 0 0 | + | 8 20 1 0 0 0 0 |
| − | 3 21 1 0 0 0 0 | + | 3 21 1 0 0 0 0 |
| − | 14 22 1 0 0 0 0 | + | 14 22 1 0 0 0 0 |
| − | 23 24 1 0 0 0 0 | + | 23 24 1 0 0 0 0 |
| − | 24 25 1 1 0 0 0 | + | 24 25 1 1 0 0 0 |
| − | 25 26 1 1 0 0 0 | + | 25 26 1 1 0 0 0 |
| − | 27 26 1 1 0 0 0 | + | 27 26 1 1 0 0 0 |
| − | 27 28 1 0 0 0 0 | + | 27 28 1 0 0 0 0 |
| − | 28 23 1 0 0 0 0 | + | 28 23 1 0 0 0 0 |
| − | 23 29 1 0 0 0 0 | + | 23 29 1 0 0 0 0 |
| − | 28 30 1 0 0 0 0 | + | 28 30 1 0 0 0 0 |
| − | 27 31 1 0 0 0 0 | + | 27 31 1 0 0 0 0 |
| − | 24 22 1 0 0 0 0 | + | 24 22 1 0 0 0 0 |
| − | 32 33 1 0 0 0 0 | + | 32 33 1 0 0 0 0 |
| − | 33 34 1 1 0 0 0 | + | 33 34 1 1 0 0 0 |
| − | 34 35 1 1 0 0 0 | + | 34 35 1 1 0 0 0 |
| − | 36 35 1 1 0 0 0 | + | 36 35 1 1 0 0 0 |
| − | 36 37 1 0 0 0 0 | + | 36 37 1 0 0 0 0 |
| − | 37 32 1 0 0 0 0 | + | 37 32 1 0 0 0 0 |
| − | 32 38 1 0 0 0 0 | + | 32 38 1 0 0 0 0 |
| − | 37 39 1 0 0 0 0 | + | 37 39 1 0 0 0 0 |
| − | 36 40 1 0 0 0 0 | + | 36 40 1 0 0 0 0 |
| − | 33 20 1 0 0 0 0 | + | 33 20 1 0 0 0 0 |
| − | 35 41 1 0 0 0 0 | + | 35 41 1 0 0 0 0 |
| − | 41 42 1 0 0 0 0 | + | 41 42 1 0 0 0 0 |
| − | 26 43 1 0 0 0 0 | + | 26 43 1 0 0 0 0 |
| − | 43 44 1 0 0 0 0 | + | 43 44 1 0 0 0 0 |
| − | 16 45 1 0 0 0 0 | + | 16 45 1 0 0 0 0 |
| − | 45 46 1 0 0 0 0 | + | 45 46 1 0 0 0 0 |
| − | M STY 1 3 SUP | + | M STY 1 3 SUP |
| − | M SLB 1 3 3 | + | M SLB 1 3 3 |
| − | M SAL 3 2 43 44 | + | M SAL 3 2 43 44 |
| − | M SBL 3 1 47 | + | M SBL 3 1 47 |
| − | M SMT 3 CH2OH | + | M SMT 3 CH2OH |
| − | M SVB 3 47 3.2004 -0.7098 | + | M SVB 3 47 3.2004 -0.7098 |
| − | M STY 1 2 SUP | + | M STY 1 2 SUP |
| − | M SLB 1 2 2 | + | M SLB 1 2 2 |
| − | M SAL 2 2 41 42 | + | M SAL 2 2 41 42 |
| − | M SBL 2 1 45 | + | M SBL 2 1 45 |
| − | M SMT 2 CH2OH | + | M SMT 2 CH2OH |
| − | M SVB 2 45 1.3029 -1.7617 | + | M SVB 2 45 1.3029 -1.7617 |
| − | M STY 1 1 SUP | + | M STY 1 1 SUP |
| − | M SLB 1 1 1 | + | M SLB 1 1 1 |
| − | M SAL 1 2 45 46 | + | M SAL 1 2 45 46 |
| − | M SBL 1 1 49 | + | M SBL 1 1 49 |
| − | M SMT 1 OCH3 | + | M SMT 1 OCH3 |
| − | M SVB 1 49 0.2729 2.1742 | + | M SVB 1 49 0.2729 2.1742 |
| − | S SKP 8 | + | S SKP 8 |
| − | ID FL5FAHGL0003 | + | ID FL5FAHGL0003 |
| − | KNApSAcK_ID C00005766 | + | KNApSAcK_ID C00005766 |
| − | NAME Laricitrin 3,5'-diglucoside | + | NAME Laricitrin 3,5'-diglucoside |
| − | CAS_RN 89345-43-7 | + | CAS_RN 89345-43-7 |
| − | FORMULA C28H32O18 | + | FORMULA C28H32O18 |
| − | EXACTMASS 656.1588642199999 | + | EXACTMASS 656.1588642199999 |
| − | AVERAGEMASS 656.54288 | + | AVERAGEMASS 656.54288 |
| − | SMILES O([C@H]1Oc(c2O)cc(C(=C(O[C@@H](C(O)5)O[C@@H]([C@H](O)C(O)5)CO)4)Oc(c3)c(C(=O)4)c(O)cc3O)cc(OC)2)[C@@H]([C@H](O)C(C1O)O)CO | + | SMILES O([C@H]1Oc(c2O)cc(C(=C(O[C@@H](C(O)5)O[C@@H]([C@H](O)C(O)5)CO)4)Oc(c3)c(C(=O)4)c(O)cc3O)cc(OC)2)[C@@H]([C@H](O)C(C1O)O)CO |
M END | M END | ||
| − | |||
Latest revision as of 09:00, 14 March 2009
Copyright: ARM project http://www.metabolome.jp/
46 50 0 0 0 0 0 0 0 0999 V2000
-3.3587 0.2950 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.3587 -0.3474 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.8024 -0.6686 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.2461 -0.3474 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.2461 0.2950 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.8024 0.6161 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.6898 -0.6686 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.1335 -0.3474 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.1335 0.2950 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.6898 0.6161 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.6898 -1.1694 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.5774 0.6160 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.0105 0.2887 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.5565 0.6160 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.5565 1.2707 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.0105 1.5981 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.5774 1.2707 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.9148 0.6160 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.4225 1.7707 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.7394 -0.7363 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.8024 -1.3107 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.1233 0.2888 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.2861 0.3775 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0
1.7650 0.0767 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
2.3487 -0.0694 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.7700 -0.5036 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
3.2911 -0.2028 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
2.7074 -0.0567 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0
1.7274 0.5272 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.9807 0.3336 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.7752 -0.0731 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.3886 -0.6744 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0
-0.1325 -0.9752 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
0.4512 -1.1213 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.8725 -1.5554 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
1.3936 -1.2547 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
0.8099 -1.1085 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0
-0.1701 -0.5247 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.0832 -0.7182 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.8871 -1.3417 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.9090 -2.0313 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.9090 -2.8562 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.0043 -0.9194 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.3529 -1.6670 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.2729 2.1742 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.7143 3.0715 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1 2 1 0 0 0 0
2 3 2 0 0 0 0
3 4 1 0 0 0 0
4 5 2 0 0 0 0
5 6 1 0 0 0 0
6 1 2 0 0 0 0
4 7 1 0 0 0 0
7 8 1 0 0 0 0
8 9 2 0 0 0 0
9 10 1 0 0 0 0
10 5 1 0 0 0 0
7 11 2 0 0 0 0
9 12 1 0 0 0 0
12 13 2 0 0 0 0
13 14 1 0 0 0 0
14 15 2 0 0 0 0
15 16 1 0 0 0 0
16 17 2 0 0 0 0
17 12 1 0 0 0 0
1 18 1 0 0 0 0
15 19 1 0 0 0 0
8 20 1 0 0 0 0
3 21 1 0 0 0 0
14 22 1 0 0 0 0
23 24 1 0 0 0 0
24 25 1 1 0 0 0
25 26 1 1 0 0 0
27 26 1 1 0 0 0
27 28 1 0 0 0 0
28 23 1 0 0 0 0
23 29 1 0 0 0 0
28 30 1 0 0 0 0
27 31 1 0 0 0 0
24 22 1 0 0 0 0
32 33 1 0 0 0 0
33 34 1 1 0 0 0
34 35 1 1 0 0 0
36 35 1 1 0 0 0
36 37 1 0 0 0 0
37 32 1 0 0 0 0
32 38 1 0 0 0 0
37 39 1 0 0 0 0
36 40 1 0 0 0 0
33 20 1 0 0 0 0
35 41 1 0 0 0 0
41 42 1 0 0 0 0
26 43 1 0 0 0 0
43 44 1 0 0 0 0
16 45 1 0 0 0 0
45 46 1 0 0 0 0
M STY 1 3 SUP
M SLB 1 3 3
M SAL 3 2 43 44
M SBL 3 1 47
M SMT 3 CH2OH
M SVB 3 47 3.2004 -0.7098
M STY 1 2 SUP
M SLB 1 2 2
M SAL 2 2 41 42
M SBL 2 1 45
M SMT 2 CH2OH
M SVB 2 45 1.3029 -1.7617
M STY 1 1 SUP
M SLB 1 1 1
M SAL 1 2 45 46
M SBL 1 1 49
M SMT 1 OCH3
M SVB 1 49 0.2729 2.1742
S SKP 8
ID FL5FAHGL0003
KNApSAcK_ID C00005766
NAME Laricitrin 3,5'-diglucoside
CAS_RN 89345-43-7
FORMULA C28H32O18
EXACTMASS 656.1588642199999
AVERAGEMASS 656.54288
SMILES O([C@H]1Oc(c2O)cc(C(=C(O[C@@H](C(O)5)O[C@@H]([C@H](O)C(O)5)CO)4)Oc(c3)c(C(=O)4)c(O)cc3O)cc(OC)2)[C@@H]([C@H](O)C(C1O)O)CO
M END