Mol:FL5FAEGL0003
From Metabolomics.JP
(Difference between revisions)
| Line 1: | Line 1: | ||
| − | + | ||
| − | + | ||
| − | Copyright: ARM project http://www.metabolome.jp/ | + | Copyright: ARM project http://www.metabolome.jp/ |
| − | 45 49 0 0 0 0 0 0 0 0999 V2000 | + | 45 49 0 0 0 0 0 0 0 0999 V2000 |
| − | -1.3151 -0.2318 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.3151 -0.2318 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.3151 -0.8742 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.3151 -0.8742 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.7588 -1.1953 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.7588 -1.1953 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.2025 -0.8742 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.2025 -0.8742 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.2025 -0.2318 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.2025 -0.2318 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.7588 0.0894 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.7588 0.0894 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.3538 -1.1953 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.3538 -1.1953 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.9101 -0.8742 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.9101 -0.8742 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.9101 -0.2318 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.9101 -0.2318 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.3538 0.0894 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.3538 0.0894 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.3538 -1.6962 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.3538 -1.6962 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.6106 0.2130 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.6106 0.2130 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.1776 -0.1143 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.1776 -0.1143 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.7445 0.2130 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.7445 0.2130 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.7445 0.8677 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.7445 0.8677 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.1776 1.1950 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.1776 1.1950 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.6106 0.8677 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.6106 0.8677 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.7588 -1.8375 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.7588 -1.8375 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.9796 0.1518 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.9796 0.1518 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.1776 1.8495 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.1776 1.8495 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.3541 -1.3182 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.3541 -1.3182 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.5868 -0.9029 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.5868 -0.9029 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.2860 -1.4239 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.2860 -1.4239 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.8644 -1.2586 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.8644 -1.2586 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.4464 -1.4239 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.4464 -1.4239 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.7473 -0.9029 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.7473 -0.9029 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.1688 -1.0682 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.1688 -1.0682 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.0281 -1.0526 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.0281 -1.0526 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.4070 -0.6556 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.4070 -0.6556 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.4711 -1.0969 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.4711 -1.0969 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.0107 0.0333 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.0107 0.0333 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.6395 -0.4566 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.6395 -0.4566 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.1050 -0.2488 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.1050 -0.2488 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.5892 -0.2432 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.5892 -0.2432 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.9640 0.1317 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.9640 0.1317 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.4316 -0.1151 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.4316 -0.1151 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.5245 -0.2633 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.5245 -0.2633 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.2181 -0.4848 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.2181 -0.4848 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.7987 -0.7629 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.7987 -0.7629 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.3514 1.2180 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.3514 1.2180 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.0658 0.8055 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.0658 0.8055 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.8100 -1.4370 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.8100 -1.4370 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.5245 -1.8495 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.5245 -1.8495 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.8301 0.6691 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.8301 0.6691 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.0332 0.4555 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.0332 0.4555 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1 2 1 0 0 0 0 | + | 1 2 1 0 0 0 0 |
| − | 2 3 2 0 0 0 0 | + | 2 3 2 0 0 0 0 |
| − | 4 5 2 0 0 0 0 | + | 4 5 2 0 0 0 0 |
| − | 5 6 1 0 0 0 0 | + | 5 6 1 0 0 0 0 |
| − | 6 1 2 0 0 0 0 | + | 6 1 2 0 0 0 0 |
| − | 4 7 1 0 0 0 0 | + | 4 7 1 0 0 0 0 |
| − | 7 8 1 0 0 0 0 | + | 7 8 1 0 0 0 0 |
| − | 8 9 2 0 0 0 0 | + | 8 9 2 0 0 0 0 |
| − | 9 10 1 0 0 0 0 | + | 9 10 1 0 0 0 0 |
| − | 10 5 1 0 0 0 0 | + | 10 5 1 0 0 0 0 |
| − | 7 11 2 0 0 0 0 | + | 7 11 2 0 0 0 0 |
| − | 9 12 1 0 0 0 0 | + | 9 12 1 0 0 0 0 |
| − | 12 13 2 0 0 0 0 | + | 12 13 2 0 0 0 0 |
| − | 13 14 1 0 0 0 0 | + | 13 14 1 0 0 0 0 |
| − | 14 15 2 0 0 0 0 | + | 14 15 2 0 0 0 0 |
| − | 15 16 1 0 0 0 0 | + | 15 16 1 0 0 0 0 |
| − | 16 17 2 0 0 0 0 | + | 16 17 2 0 0 0 0 |
| − | 17 12 1 0 0 0 0 | + | 17 12 1 0 0 0 0 |
| − | 3 18 1 0 0 0 0 | + | 3 18 1 0 0 0 0 |
| − | 4 3 1 0 0 0 0 | + | 4 3 1 0 0 0 0 |
| − | 1 19 1 0 0 0 0 | + | 1 19 1 0 0 0 0 |
| − | 16 20 1 0 0 0 0 | + | 16 20 1 0 0 0 0 |
| − | 21 8 1 0 0 0 0 | + | 21 8 1 0 0 0 0 |
| − | 22 23 1 0 0 0 0 | + | 22 23 1 0 0 0 0 |
| − | 23 24 1 1 0 0 0 | + | 23 24 1 1 0 0 0 |
| − | 24 25 1 1 0 0 0 | + | 24 25 1 1 0 0 0 |
| − | 26 25 1 1 0 0 0 | + | 26 25 1 1 0 0 0 |
| − | 26 27 1 0 0 0 0 | + | 26 27 1 0 0 0 0 |
| − | 27 22 1 0 0 0 0 | + | 27 22 1 0 0 0 0 |
| − | 22 28 1 0 0 0 0 | + | 22 28 1 0 0 0 0 |
| − | 27 29 1 0 0 0 0 | + | 27 29 1 0 0 0 0 |
| − | 26 30 1 0 0 0 0 | + | 26 30 1 0 0 0 0 |
| − | 23 21 1 0 0 0 0 | + | 23 21 1 0 0 0 0 |
| − | 31 32 1 1 0 0 0 | + | 31 32 1 1 0 0 0 |
| − | 32 33 1 1 0 0 0 | + | 32 33 1 1 0 0 0 |
| − | 34 33 1 1 0 0 0 | + | 34 33 1 1 0 0 0 |
| − | 34 35 1 0 0 0 0 | + | 34 35 1 0 0 0 0 |
| − | 35 36 1 0 0 0 0 | + | 35 36 1 0 0 0 0 |
| − | 36 31 1 0 0 0 0 | + | 36 31 1 0 0 0 0 |
| − | 31 37 1 0 0 0 0 | + | 31 37 1 0 0 0 0 |
| − | 32 38 1 0 0 0 0 | + | 32 38 1 0 0 0 0 |
| − | 33 39 1 0 0 0 0 | + | 33 39 1 0 0 0 0 |
| − | 34 19 1 0 0 0 0 | + | 34 19 1 0 0 0 0 |
| − | 15 40 1 0 0 0 0 | + | 15 40 1 0 0 0 0 |
| − | 40 41 1 0 0 0 0 | + | 40 41 1 0 0 0 0 |
| − | 25 42 1 0 0 0 0 | + | 25 42 1 0 0 0 0 |
| − | 42 43 1 0 0 0 0 | + | 42 43 1 0 0 0 0 |
| − | 36 44 1 0 0 0 0 | + | 36 44 1 0 0 0 0 |
| − | 44 45 1 0 0 0 0 | + | 44 45 1 0 0 0 0 |
| − | M STY 1 1 SUP | + | M STY 1 1 SUP |
| − | M SLB 1 1 1 | + | M SLB 1 1 1 |
| − | M SAL 1 2 40 41 | + | M SAL 1 2 40 41 |
| − | M SBL 1 1 44 | + | M SBL 1 1 44 |
| − | M SMT 1 OCH3 | + | M SMT 1 OCH3 |
| − | M SBV 1 44 -4.3754 8.3273 | + | M SBV 1 44 -4.3754 8.3273 |
| − | M STY 1 2 SUP | + | M STY 1 2 SUP |
| − | M SLB 1 2 2 | + | M SLB 1 2 2 |
| − | M SAL 2 2 42 43 | + | M SAL 2 2 42 43 |
| − | M SBL 2 1 46 | + | M SBL 2 1 46 |
| − | M SMT 2 CH2OH | + | M SMT 2 CH2OH |
| − | M SBV 2 46 -4.6186 7.9639 | + | M SBV 2 46 -4.6186 7.9639 |
| − | M STY 1 3 SUP | + | M STY 1 3 SUP |
| − | M SLB 1 3 3 | + | M SLB 1 3 3 |
| − | M SAL 3 2 44 45 | + | M SAL 3 2 44 45 |
| − | M SBL 3 1 48 | + | M SBL 3 1 48 |
| − | M SMT 3 ^CH2OH | + | M SMT 3 ^CH2OH |
| − | M SBV 3 48 -5.3807 8.7611 | + | M SBV 3 48 -5.3807 8.7611 |
| − | S SKP 8 | + | S SKP 8 |
| − | ID FL5FAEGL0003 | + | ID FL5FAEGL0003 |
| − | KNApSAcK_ID C00005597 | + | KNApSAcK_ID C00005597 |
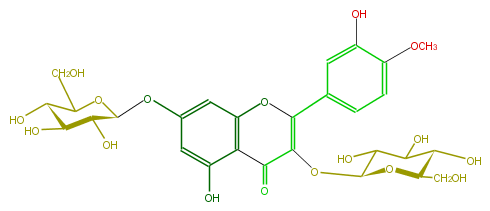
| − | NAME Tamarixetin 3,7-diglucoside;Quercetin 4'-methyl ether 3,7-diglucoside;3,7-Bis(beta-D-glucopyranosyloxy)-5-hydroxy-2-(3-hydroxy-4-methoxyphenyl)-4H-1-benzopyran-4-one | + | NAME Tamarixetin 3,7-diglucoside;Quercetin 4'-methyl ether 3,7-diglucoside;3,7-Bis(beta-D-glucopyranosyloxy)-5-hydroxy-2-(3-hydroxy-4-methoxyphenyl)-4H-1-benzopyran-4-one |
| − | CAS_RN 165460-84-4 | + | CAS_RN 165460-84-4 |
| − | FORMULA C28H32O17 | + | FORMULA C28H32O17 |
| − | EXACTMASS 640.163949598 | + | EXACTMASS 640.163949598 |
| − | AVERAGEMASS 640.54348 | + | AVERAGEMASS 640.54348 |
| − | SMILES c(c1C(=C4OC(O5)C(C(C(O)C5CO)O)O)Oc(c2)c(C4=O)c(cc2OC(O3)C(O)C(O)C(O)C(CO)3)O)cc(OC)c(O)c1 | + | SMILES c(c1C(=C4OC(O5)C(C(C(O)C5CO)O)O)Oc(c2)c(C4=O)c(cc2OC(O3)C(O)C(O)C(O)C(CO)3)O)cc(OC)c(O)c1 |
M END | M END | ||
| − | |||
Latest revision as of 09:00, 14 March 2009
Copyright: ARM project http://www.metabolome.jp/
45 49 0 0 0 0 0 0 0 0999 V2000
-1.3151 -0.2318 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.3151 -0.8742 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.7588 -1.1953 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.2025 -0.8742 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.2025 -0.2318 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.7588 0.0894 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.3538 -1.1953 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.9101 -0.8742 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.9101 -0.2318 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.3538 0.0894 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.3538 -1.6962 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.6106 0.2130 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.1776 -0.1143 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.7445 0.2130 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.7445 0.8677 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.1776 1.1950 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.6106 0.8677 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.7588 -1.8375 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.9796 0.1518 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.1776 1.8495 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.3541 -1.3182 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.5868 -0.9029 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.2860 -1.4239 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.8644 -1.2586 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.4464 -1.4239 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.7473 -0.9029 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.1688 -1.0682 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.0281 -1.0526 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.4070 -0.6556 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
4.4711 -1.0969 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.0107 0.0333 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.6395 -0.4566 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.1050 -0.2488 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.5892 -0.2432 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.9640 0.1317 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.4316 -0.1151 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.5245 -0.2633 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.2181 -0.4848 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.7987 -0.7629 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.3514 1.2180 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
4.0658 0.8055 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.8100 -1.4370 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.5245 -1.8495 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.8301 0.6691 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.0332 0.4555 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1 2 1 0 0 0 0
2 3 2 0 0 0 0
4 5 2 0 0 0 0
5 6 1 0 0 0 0
6 1 2 0 0 0 0
4 7 1 0 0 0 0
7 8 1 0 0 0 0
8 9 2 0 0 0 0
9 10 1 0 0 0 0
10 5 1 0 0 0 0
7 11 2 0 0 0 0
9 12 1 0 0 0 0
12 13 2 0 0 0 0
13 14 1 0 0 0 0
14 15 2 0 0 0 0
15 16 1 0 0 0 0
16 17 2 0 0 0 0
17 12 1 0 0 0 0
3 18 1 0 0 0 0
4 3 1 0 0 0 0
1 19 1 0 0 0 0
16 20 1 0 0 0 0
21 8 1 0 0 0 0
22 23 1 0 0 0 0
23 24 1 1 0 0 0
24 25 1 1 0 0 0
26 25 1 1 0 0 0
26 27 1 0 0 0 0
27 22 1 0 0 0 0
22 28 1 0 0 0 0
27 29 1 0 0 0 0
26 30 1 0 0 0 0
23 21 1 0 0 0 0
31 32 1 1 0 0 0
32 33 1 1 0 0 0
34 33 1 1 0 0 0
34 35 1 0 0 0 0
35 36 1 0 0 0 0
36 31 1 0 0 0 0
31 37 1 0 0 0 0
32 38 1 0 0 0 0
33 39 1 0 0 0 0
34 19 1 0 0 0 0
15 40 1 0 0 0 0
40 41 1 0 0 0 0
25 42 1 0 0 0 0
42 43 1 0 0 0 0
36 44 1 0 0 0 0
44 45 1 0 0 0 0
M STY 1 1 SUP
M SLB 1 1 1
M SAL 1 2 40 41
M SBL 1 1 44
M SMT 1 OCH3
M SBV 1 44 -4.3754 8.3273
M STY 1 2 SUP
M SLB 1 2 2
M SAL 2 2 42 43
M SBL 2 1 46
M SMT 2 CH2OH
M SBV 2 46 -4.6186 7.9639
M STY 1 3 SUP
M SLB 1 3 3
M SAL 3 2 44 45
M SBL 3 1 48
M SMT 3 ^CH2OH
M SBV 3 48 -5.3807 8.7611
S SKP 8
ID FL5FAEGL0003
KNApSAcK_ID C00005597
NAME Tamarixetin 3,7-diglucoside;Quercetin 4'-methyl ether 3,7-diglucoside;3,7-Bis(beta-D-glucopyranosyloxy)-5-hydroxy-2-(3-hydroxy-4-methoxyphenyl)-4H-1-benzopyran-4-one
CAS_RN 165460-84-4
FORMULA C28H32O17
EXACTMASS 640.163949598
AVERAGEMASS 640.54348
SMILES c(c1C(=C4OC(O5)C(C(C(O)C5CO)O)O)Oc(c2)c(C4=O)c(cc2OC(O3)C(O)C(O)C(O)C(CO)3)O)cc(OC)c(O)c1
M END