Mol:FL5FADGL0024
From Metabolomics.JP
(Difference between revisions)
| Line 1: | Line 1: | ||
| − | + | ||
| − | + | ||
| − | Copyright: ARM project http://www.metabolome.jp/ | + | Copyright: ARM project http://www.metabolome.jp/ |
| − | 56 61 0 0 0 0 0 0 0 0999 V2000 | + | 56 61 0 0 0 0 0 0 0 0999 V2000 |
| − | -1.4417 1.2233 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.4417 1.2233 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.4417 0.5809 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.4417 0.5809 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.8854 0.2597 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.8854 0.2597 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.3291 0.5809 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.3291 0.5809 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.3291 1.2233 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.3291 1.2233 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.8854 1.5445 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.8854 1.5445 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.2272 0.2597 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.2272 0.2597 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.7835 0.5809 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.7835 0.5809 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.7835 1.2233 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.7835 1.2233 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.2272 1.5445 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.2272 1.5445 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.2272 -0.2411 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.2272 -0.2411 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.3396 1.5443 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.3396 1.5443 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.9066 1.2170 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.9066 1.2170 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.4736 1.5443 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.4736 1.5443 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.4736 2.1990 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.4736 2.1990 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.9066 2.5264 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.9066 2.5264 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.3396 2.1990 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.3396 2.1990 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.9978 1.5443 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.9978 1.5443 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.1777 0.1920 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.1777 0.1920 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.8854 -0.3824 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.8854 -0.3824 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.3396 2.6990 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.3396 2.6990 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.5683 -0.9257 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 | + | 1.5683 -0.9257 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 |
| − | 0.9855 -0.5893 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | 0.9855 -0.5893 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | 1.1704 -1.2363 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.1704 -1.2363 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.9855 -1.8873 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | 0.9855 -1.8873 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | 1.5683 -2.2238 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | 1.5683 -2.2238 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | 1.3834 -1.5767 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 | + | 1.3834 -1.5767 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 |
| − | 1.4008 -0.3008 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.4008 -0.3008 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.8146 -1.6923 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.8146 -1.6923 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.9041 -3.1657 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.9041 -3.1657 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.3345 0.4589 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0 | + | 2.3345 0.4589 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0 |
| − | 2.7769 -0.1250 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | 2.7769 -0.1250 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | 3.4139 0.1227 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | 3.4139 0.1227 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | 4.0287 0.1293 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | 4.0287 0.1293 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | 3.5820 0.5761 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 | + | 3.5820 0.5761 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 |
| − | 3.0246 0.2820 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.0246 0.2820 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.0872 -0.1586 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.0872 -0.1586 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.7790 -0.4901 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.7790 -0.4901 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 5.0199 -0.0029 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 5.0199 -0.0029 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.3055 1.1839 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0 | + | -4.3055 1.1839 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0 |
| − | -3.9001 0.6487 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | -3.9001 0.6487 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | -3.3163 0.8758 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | -3.3163 0.8758 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | -2.7078 0.8691 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | -2.7078 0.8691 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | -3.1623 1.2913 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.1623 1.2913 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.7587 1.0772 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 | + | -3.7587 1.0772 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 |
| − | -4.8269 1.2296 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.8269 1.2296 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.2828 0.1450 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.2828 0.1450 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.9818 0.3142 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.9818 0.3142 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.1900 3.1025 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.1900 3.1025 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.6314 3.9998 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.6314 3.9998 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.8903 0.8140 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.8903 0.8140 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.7572 1.8051 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.7572 1.8051 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.4672 -2.7425 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.4672 -2.7425 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.5326 -2.7212 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.5326 -2.7212 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.9793 1.7832 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.9793 1.7832 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.9550 2.0022 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.9550 2.0022 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1 2 1 0 0 0 0 | + | 1 2 1 0 0 0 0 |
| − | 2 3 2 0 0 0 0 | + | 2 3 2 0 0 0 0 |
| − | 3 4 1 0 0 0 0 | + | 3 4 1 0 0 0 0 |
| − | 4 5 2 0 0 0 0 | + | 4 5 2 0 0 0 0 |
| − | 5 6 1 0 0 0 0 | + | 5 6 1 0 0 0 0 |
| − | 6 1 2 0 0 0 0 | + | 6 1 2 0 0 0 0 |
| − | 4 7 1 0 0 0 0 | + | 4 7 1 0 0 0 0 |
| − | 7 8 1 0 0 0 0 | + | 7 8 1 0 0 0 0 |
| − | 8 9 2 0 0 0 0 | + | 8 9 2 0 0 0 0 |
| − | 9 10 1 0 0 0 0 | + | 9 10 1 0 0 0 0 |
| − | 10 5 1 0 0 0 0 | + | 10 5 1 0 0 0 0 |
| − | 7 11 2 0 0 0 0 | + | 7 11 2 0 0 0 0 |
| − | 9 12 1 0 0 0 0 | + | 9 12 1 0 0 0 0 |
| − | 12 13 2 0 0 0 0 | + | 12 13 2 0 0 0 0 |
| − | 13 14 1 0 0 0 0 | + | 13 14 1 0 0 0 0 |
| − | 14 15 2 0 0 0 0 | + | 14 15 2 0 0 0 0 |
| − | 15 16 1 0 0 0 0 | + | 15 16 1 0 0 0 0 |
| − | 16 17 2 0 0 0 0 | + | 16 17 2 0 0 0 0 |
| − | 17 12 1 0 0 0 0 | + | 17 12 1 0 0 0 0 |
| − | 1 18 1 0 0 0 0 | + | 1 18 1 0 0 0 0 |
| − | 8 19 1 0 0 0 0 | + | 8 19 1 0 0 0 0 |
| − | 3 20 1 0 0 0 0 | + | 3 20 1 0 0 0 0 |
| − | 21 15 1 0 0 0 0 | + | 21 15 1 0 0 0 0 |
| − | 22 23 1 0 0 0 0 | + | 22 23 1 0 0 0 0 |
| − | 23 24 1 1 0 0 0 | + | 23 24 1 1 0 0 0 |
| − | 24 25 1 1 0 0 0 | + | 24 25 1 1 0 0 0 |
| − | 26 25 1 1 0 0 0 | + | 26 25 1 1 0 0 0 |
| − | 26 27 1 0 0 0 0 | + | 26 27 1 0 0 0 0 |
| − | 27 22 1 0 0 0 0 | + | 27 22 1 0 0 0 0 |
| − | 22 28 1 0 0 0 0 | + | 22 28 1 0 0 0 0 |
| − | 27 29 1 0 0 0 0 | + | 27 29 1 0 0 0 0 |
| − | 26 30 1 0 0 0 0 | + | 26 30 1 0 0 0 0 |
| − | 23 19 1 0 0 0 0 | + | 23 19 1 0 0 0 0 |
| − | 31 32 1 1 0 0 0 | + | 31 32 1 1 0 0 0 |
| − | 32 33 1 1 0 0 0 | + | 32 33 1 1 0 0 0 |
| − | 34 33 1 1 0 0 0 | + | 34 33 1 1 0 0 0 |
| − | 34 35 1 0 0 0 0 | + | 34 35 1 0 0 0 0 |
| − | 35 36 1 0 0 0 0 | + | 35 36 1 0 0 0 0 |
| − | 36 31 1 0 0 0 0 | + | 36 31 1 0 0 0 0 |
| − | 32 37 1 0 0 0 0 | + | 32 37 1 0 0 0 0 |
| − | 33 38 1 0 0 0 0 | + | 33 38 1 0 0 0 0 |
| − | 31 28 1 0 0 0 0 | + | 31 28 1 0 0 0 0 |
| − | 34 39 1 0 0 0 0 | + | 34 39 1 0 0 0 0 |
| − | 40 41 1 1 0 0 0 | + | 40 41 1 1 0 0 0 |
| − | 41 42 1 1 0 0 0 | + | 41 42 1 1 0 0 0 |
| − | 43 42 1 1 0 0 0 | + | 43 42 1 1 0 0 0 |
| − | 43 44 1 0 0 0 0 | + | 43 44 1 0 0 0 0 |
| − | 44 45 1 0 0 0 0 | + | 44 45 1 0 0 0 0 |
| − | 45 40 1 0 0 0 0 | + | 45 40 1 0 0 0 0 |
| − | 40 46 1 0 0 0 0 | + | 40 46 1 0 0 0 0 |
| − | 41 47 1 0 0 0 0 | + | 41 47 1 0 0 0 0 |
| − | 42 48 1 0 0 0 0 | + | 42 48 1 0 0 0 0 |
| − | 43 18 1 0 0 0 0 | + | 43 18 1 0 0 0 0 |
| − | 16 49 1 0 0 0 0 | + | 16 49 1 0 0 0 0 |
| − | 49 50 1 0 0 0 0 | + | 49 50 1 0 0 0 0 |
| − | 35 51 1 0 0 0 0 | + | 35 51 1 0 0 0 0 |
| − | 51 52 1 0 0 0 0 | + | 51 52 1 0 0 0 0 |
| − | 25 53 1 0 0 0 0 | + | 25 53 1 0 0 0 0 |
| − | 53 54 1 0 0 0 0 | + | 53 54 1 0 0 0 0 |
| − | 45 55 1 0 0 0 0 | + | 45 55 1 0 0 0 0 |
| − | 55 56 1 0 0 0 0 | + | 55 56 1 0 0 0 0 |
| − | M STY 1 4 SUP | + | M STY 1 4 SUP |
| − | M SLB 1 4 4 | + | M SLB 1 4 4 |
| − | M SAL 4 2 55 56 | + | M SAL 4 2 55 56 |
| − | M SBL 4 1 60 | + | M SBL 4 1 60 |
| − | M SMT 4 CH2OH | + | M SMT 4 CH2OH |
| − | M SVB 4 60 -3.9793 1.7832 | + | M SVB 4 60 -3.9793 1.7832 |
| − | M STY 1 3 SUP | + | M STY 1 3 SUP |
| − | M SLB 1 3 3 | + | M SLB 1 3 3 |
| − | M SAL 3 2 53 54 | + | M SAL 3 2 53 54 |
| − | M SBL 3 1 58 | + | M SBL 3 1 58 |
| − | M SMT 3 CH2OH | + | M SMT 3 CH2OH |
| − | M SVB 3 58 1.3726 -2.5191 | + | M SVB 3 58 1.3726 -2.5191 |
| − | M STY 1 2 SUP | + | M STY 1 2 SUP |
| − | M SLB 1 2 2 | + | M SLB 1 2 2 |
| − | M SAL 2 2 51 52 | + | M SAL 2 2 51 52 |
| − | M SBL 2 1 56 | + | M SBL 2 1 56 |
| − | M SMT 2 CH2OH | + | M SMT 2 CH2OH |
| − | M SVB 2 56 3.962 0.6614 | + | M SVB 2 56 3.962 0.6614 |
| − | M STY 1 1 SUP | + | M STY 1 1 SUP |
| − | M SLB 1 1 1 | + | M SLB 1 1 1 |
| − | M SAL 1 2 49 50 | + | M SAL 1 2 49 50 |
| − | M SBL 1 1 54 | + | M SBL 1 1 54 |
| − | M SMT 1 OCH3 | + | M SMT 1 OCH3 |
| − | M SVB 1 54 2.19 3.1025 | + | M SVB 1 54 2.19 3.1025 |
| − | S SKP 8 | + | S SKP 8 |
| − | ID FL5FADGL0024 | + | ID FL5FADGL0024 |
| − | KNApSAcK_ID C00005579 | + | KNApSAcK_ID C00005579 |
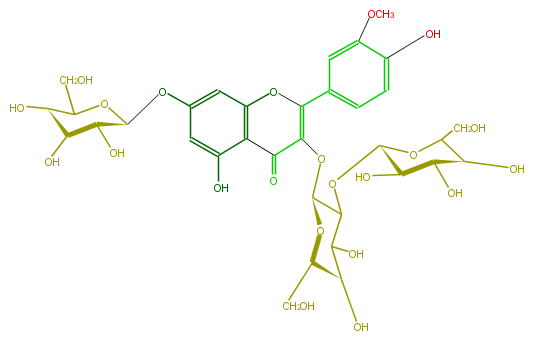
| − | NAME Isorhamnetin 3-sophoroside-7-glucoside | + | NAME Isorhamnetin 3-sophoroside-7-glucoside |
| − | CAS_RN 17331-29-2 | + | CAS_RN 17331-29-2 |
| − | FORMULA C34H42O22 | + | FORMULA C34H42O22 |
| − | EXACTMASS 802.216773028 | + | EXACTMASS 802.216773028 |
| − | AVERAGEMASS 802.68408 | + | AVERAGEMASS 802.68408 |
| − | SMILES C(C([C@H]6O)O[C@@H]([C@@H]([C@@H]6O)O)OC([C@H](OC(=C2c(c5)cc(c(c5)O)OC)C(c(c4O)c(cc(c4)O[C@@H]([C@@H](O)3)OC([C@@H]([C@H](O)3)O)CO)O2)=O)1)C([C@H]([C@@H](CO)O1)O)O)O | + | SMILES C(C([C@H]6O)O[C@@H]([C@@H]([C@@H]6O)O)OC([C@H](OC(=C2c(c5)cc(c(c5)O)OC)C(c(c4O)c(cc(c4)O[C@@H]([C@@H](O)3)OC([C@@H]([C@H](O)3)O)CO)O2)=O)1)C([C@H]([C@@H](CO)O1)O)O)O |
M END | M END | ||
| − | |||
Latest revision as of 09:00, 14 March 2009
Copyright: ARM project http://www.metabolome.jp/
56 61 0 0 0 0 0 0 0 0999 V2000
-1.4417 1.2233 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.4417 0.5809 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.8854 0.2597 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.3291 0.5809 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.3291 1.2233 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.8854 1.5445 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.2272 0.2597 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.7835 0.5809 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.7835 1.2233 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.2272 1.5445 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.2272 -0.2411 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.3396 1.5443 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.9066 1.2170 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.4736 1.5443 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.4736 2.1990 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.9066 2.5264 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.3396 2.1990 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.9978 1.5443 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.1777 0.1920 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.8854 -0.3824 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.3396 2.6990 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.5683 -0.9257 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0
0.9855 -0.5893 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
1.1704 -1.2363 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.9855 -1.8873 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
1.5683 -2.2238 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
1.3834 -1.5767 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0
1.4008 -0.3008 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.8146 -1.6923 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.9041 -3.1657 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.3345 0.4589 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0
2.7769 -0.1250 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
3.4139 0.1227 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
4.0287 0.1293 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
3.5820 0.5761 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0
3.0246 0.2820 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.0872 -0.1586 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.7790 -0.4901 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
5.0199 -0.0029 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.3055 1.1839 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0
-3.9001 0.6487 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
-3.3163 0.8758 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
-2.7078 0.8691 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
-3.1623 1.2913 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.7587 1.0772 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0
-4.8269 1.2296 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.2828 0.1450 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.9818 0.3142 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.1900 3.1025 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.6314 3.9998 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.8903 0.8140 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.7572 1.8051 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.4672 -2.7425 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.5326 -2.7212 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.9793 1.7832 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.9550 2.0022 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1 2 1 0 0 0 0
2 3 2 0 0 0 0
3 4 1 0 0 0 0
4 5 2 0 0 0 0
5 6 1 0 0 0 0
6 1 2 0 0 0 0
4 7 1 0 0 0 0
7 8 1 0 0 0 0
8 9 2 0 0 0 0
9 10 1 0 0 0 0
10 5 1 0 0 0 0
7 11 2 0 0 0 0
9 12 1 0 0 0 0
12 13 2 0 0 0 0
13 14 1 0 0 0 0
14 15 2 0 0 0 0
15 16 1 0 0 0 0
16 17 2 0 0 0 0
17 12 1 0 0 0 0
1 18 1 0 0 0 0
8 19 1 0 0 0 0
3 20 1 0 0 0 0
21 15 1 0 0 0 0
22 23 1 0 0 0 0
23 24 1 1 0 0 0
24 25 1 1 0 0 0
26 25 1 1 0 0 0
26 27 1 0 0 0 0
27 22 1 0 0 0 0
22 28 1 0 0 0 0
27 29 1 0 0 0 0
26 30 1 0 0 0 0
23 19 1 0 0 0 0
31 32 1 1 0 0 0
32 33 1 1 0 0 0
34 33 1 1 0 0 0
34 35 1 0 0 0 0
35 36 1 0 0 0 0
36 31 1 0 0 0 0
32 37 1 0 0 0 0
33 38 1 0 0 0 0
31 28 1 0 0 0 0
34 39 1 0 0 0 0
40 41 1 1 0 0 0
41 42 1 1 0 0 0
43 42 1 1 0 0 0
43 44 1 0 0 0 0
44 45 1 0 0 0 0
45 40 1 0 0 0 0
40 46 1 0 0 0 0
41 47 1 0 0 0 0
42 48 1 0 0 0 0
43 18 1 0 0 0 0
16 49 1 0 0 0 0
49 50 1 0 0 0 0
35 51 1 0 0 0 0
51 52 1 0 0 0 0
25 53 1 0 0 0 0
53 54 1 0 0 0 0
45 55 1 0 0 0 0
55 56 1 0 0 0 0
M STY 1 4 SUP
M SLB 1 4 4
M SAL 4 2 55 56
M SBL 4 1 60
M SMT 4 CH2OH
M SVB 4 60 -3.9793 1.7832
M STY 1 3 SUP
M SLB 1 3 3
M SAL 3 2 53 54
M SBL 3 1 58
M SMT 3 CH2OH
M SVB 3 58 1.3726 -2.5191
M STY 1 2 SUP
M SLB 1 2 2
M SAL 2 2 51 52
M SBL 2 1 56
M SMT 2 CH2OH
M SVB 2 56 3.962 0.6614
M STY 1 1 SUP
M SLB 1 1 1
M SAL 1 2 49 50
M SBL 1 1 54
M SMT 1 OCH3
M SVB 1 54 2.19 3.1025
S SKP 8
ID FL5FADGL0024
KNApSAcK_ID C00005579
NAME Isorhamnetin 3-sophoroside-7-glucoside
CAS_RN 17331-29-2
FORMULA C34H42O22
EXACTMASS 802.216773028
AVERAGEMASS 802.68408
SMILES C(C([C@H]6O)O[C@@H]([C@@H]([C@@H]6O)O)OC([C@H](OC(=C2c(c5)cc(c(c5)O)OC)C(c(c4O)c(cc(c4)O[C@@H]([C@@H](O)3)OC([C@@H]([C@H](O)3)O)CO)O2)=O)1)C([C@H]([C@@H](CO)O1)O)O)O
M END