Mol:FL5FADGL0015
From Metabolomics.JP
(Difference between revisions)
| Line 1: | Line 1: | ||
| − | + | ||
| − | + | ||
| − | Copyright: ARM project http://www.metabolome.jp/ | + | Copyright: ARM project http://www.metabolome.jp/ |
| − | 44 48 0 0 0 0 0 0 0 0999 V2000 | + | 44 48 0 0 0 0 0 0 0 0999 V2000 |
| − | -1.6648 0.1873 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.6648 0.1873 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.6648 -0.6381 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.6648 -0.6381 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.9500 -1.0506 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.9500 -1.0506 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.2351 -0.6381 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.2351 -0.6381 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.2352 0.1874 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.2352 0.1874 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.9500 0.6001 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.9500 0.6001 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.4797 -1.0507 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.4797 -1.0507 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.1944 -0.6381 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.1944 -0.6381 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.1944 0.1874 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.1944 0.1874 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.4797 0.6001 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.4797 0.6001 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.4798 -1.6942 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.4798 -1.6942 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.0946 0.7589 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.0946 0.7589 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.8231 0.3383 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.8231 0.3383 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.5515 0.7590 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.5515 0.7590 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.5516 1.6002 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.5516 1.6002 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.8231 2.0208 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.8231 2.0208 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.0946 1.6001 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.0946 1.6001 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.9499 -1.8758 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.9499 -1.8758 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.2326 2.0763 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.2326 2.0763 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.5186 0.6804 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.5186 0.6804 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.0344 -1.1686 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.0344 -1.1686 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.1385 -1.4595 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.1385 -1.4595 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.4504 -1.7804 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.4504 -1.7804 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.1680 -2.0281 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.1680 -2.0281 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.6498 -2.6202 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.6498 -2.6202 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.3380 -2.2994 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.3380 -2.2994 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.6204 -2.0516 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.6204 -2.0516 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.5245 -1.0656 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.5245 -1.0656 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.4413 -1.9019 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.4413 -1.9019 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.7584 -2.5862 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.7584 -2.5862 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.9534 0.4528 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.9534 0.4528 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.1858 0.3173 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.1858 0.3173 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.6434 0.9483 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.6434 0.9483 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.6999 1.7299 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.6999 1.7299 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.4674 1.8653 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.4674 1.8653 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.0100 1.2343 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.0100 1.2343 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.4582 2.4822 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.4582 2.4822 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.8603 2.2041 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.8603 2.2041 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.7584 1.3431 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.7584 1.3431 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.5384 0.2573 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.5384 0.2573 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.2403 -2.9107 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.2403 -2.9107 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.6882 -3.8715 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.6882 -3.8715 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.8174 2.7185 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.8174 2.7185 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.3846 3.8715 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.3846 3.8715 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1 2 1 0 0 0 0 | + | 1 2 1 0 0 0 0 |
| − | 2 3 2 0 0 0 0 | + | 2 3 2 0 0 0 0 |
| − | 4 5 2 0 0 0 0 | + | 4 5 2 0 0 0 0 |
| − | 5 6 1 0 0 0 0 | + | 5 6 1 0 0 0 0 |
| − | 6 1 2 0 0 0 0 | + | 6 1 2 0 0 0 0 |
| − | 4 7 1 0 0 0 0 | + | 4 7 1 0 0 0 0 |
| − | 7 8 1 0 0 0 0 | + | 7 8 1 0 0 0 0 |
| − | 8 9 2 0 0 0 0 | + | 8 9 2 0 0 0 0 |
| − | 9 10 1 0 0 0 0 | + | 9 10 1 0 0 0 0 |
| − | 10 5 1 0 0 0 0 | + | 10 5 1 0 0 0 0 |
| − | 7 11 2 0 0 0 0 | + | 7 11 2 0 0 0 0 |
| − | 9 12 1 0 0 0 0 | + | 9 12 1 0 0 0 0 |
| − | 12 13 2 0 0 0 0 | + | 12 13 2 0 0 0 0 |
| − | 13 14 1 0 0 0 0 | + | 13 14 1 0 0 0 0 |
| − | 14 15 2 0 0 0 0 | + | 14 15 2 0 0 0 0 |
| − | 15 16 1 0 0 0 0 | + | 15 16 1 0 0 0 0 |
| − | 16 17 2 0 0 0 0 | + | 16 17 2 0 0 0 0 |
| − | 17 12 1 0 0 0 0 | + | 17 12 1 0 0 0 0 |
| − | 3 18 1 0 0 0 0 | + | 3 18 1 0 0 0 0 |
| − | 19 15 1 0 0 0 0 | + | 19 15 1 0 0 0 0 |
| − | 4 3 1 0 0 0 0 | + | 4 3 1 0 0 0 0 |
| − | 1 20 1 0 0 0 0 | + | 1 20 1 0 0 0 0 |
| − | 21 8 1 0 0 0 0 | + | 21 8 1 0 0 0 0 |
| − | 22 23 1 0 0 0 0 | + | 22 23 1 0 0 0 0 |
| − | 23 24 1 1 0 0 0 | + | 23 24 1 1 0 0 0 |
| − | 24 25 1 1 0 0 0 | + | 24 25 1 1 0 0 0 |
| − | 26 25 1 1 0 0 0 | + | 26 25 1 1 0 0 0 |
| − | 26 27 1 0 0 0 0 | + | 26 27 1 0 0 0 0 |
| − | 27 22 1 0 0 0 0 | + | 27 22 1 0 0 0 0 |
| − | 22 28 1 0 0 0 0 | + | 22 28 1 0 0 0 0 |
| − | 27 29 1 0 0 0 0 | + | 27 29 1 0 0 0 0 |
| − | 26 30 1 0 0 0 0 | + | 26 30 1 0 0 0 0 |
| − | 21 23 1 0 0 0 0 | + | 21 23 1 0 0 0 0 |
| − | 32 31 1 1 0 0 0 | + | 32 31 1 1 0 0 0 |
| − | 32 33 1 0 0 0 0 | + | 32 33 1 0 0 0 0 |
| − | 33 34 1 0 0 0 0 | + | 33 34 1 0 0 0 0 |
| − | 34 35 1 0 0 0 0 | + | 34 35 1 0 0 0 0 |
| − | 35 36 1 1 0 0 0 | + | 35 36 1 1 0 0 0 |
| − | 36 31 1 1 0 0 0 | + | 36 31 1 1 0 0 0 |
| − | 35 37 1 0 0 0 0 | + | 35 37 1 0 0 0 0 |
| − | 34 38 1 0 0 0 0 | + | 34 38 1 0 0 0 0 |
| − | 36 39 1 0 0 0 0 | + | 36 39 1 0 0 0 0 |
| − | 31 40 1 0 0 0 0 | + | 31 40 1 0 0 0 0 |
| − | 32 20 1 0 0 0 0 | + | 32 20 1 0 0 0 0 |
| − | 41 42 1 0 0 0 0 | + | 41 42 1 0 0 0 0 |
| − | 25 41 1 0 0 0 0 | + | 25 41 1 0 0 0 0 |
| − | 43 44 1 0 0 0 0 | + | 43 44 1 0 0 0 0 |
| − | 16 43 1 0 0 0 0 | + | 16 43 1 0 0 0 0 |
| − | M STY 1 1 SUP | + | M STY 1 1 SUP |
| − | M SLB 1 1 1 | + | M SLB 1 1 1 |
| − | M SAL 1 2 41 42 | + | M SAL 1 2 41 42 |
| − | M SBL 1 1 46 | + | M SBL 1 1 46 |
| − | M SMT 1 CH2OH | + | M SMT 1 CH2OH |
| − | M SBV 1 46 -0.5904 0.2905 | + | M SBV 1 46 -0.5904 0.2905 |
| − | M STY 1 2 SUP | + | M STY 1 2 SUP |
| − | M SLB 1 2 2 | + | M SLB 1 2 2 |
| − | M SAL 2 2 43 44 | + | M SAL 2 2 43 44 |
| − | M SBL 2 1 48 | + | M SBL 2 1 48 |
| − | M SMT 2 OCH3 | + | M SMT 2 OCH3 |
| − | M SBV 2 48 0.0057 -0.6976 | + | M SBV 2 48 0.0057 -0.6976 |
| − | S SKP 5 | + | S SKP 5 |
| − | ID FL5FADGL0015 | + | ID FL5FADGL0015 |
| − | FORMULA C28H32O16 | + | FORMULA C28H32O16 |
| − | EXACTMASS 624.1690349759999 | + | EXACTMASS 624.1690349759999 |
| − | AVERAGEMASS 624.54408 | + | AVERAGEMASS 624.54408 |
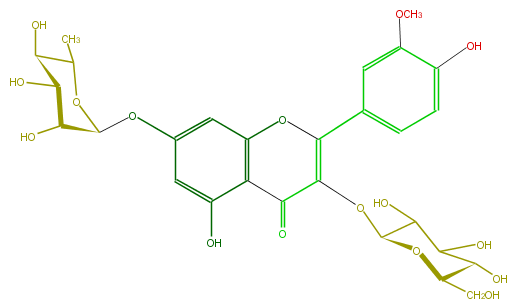
| − | SMILES OC(C1Oc(c5)cc(c3c5O)OC(c(c4)cc(OC)c(O)c4)=C(C3=O)OC(O2)C(C(C(O)C2CO)O)O)C(C(C(O1)C)O)O | + | SMILES OC(C1Oc(c5)cc(c3c5O)OC(c(c4)cc(OC)c(O)c4)=C(C3=O)OC(O2)C(C(C(O)C2CO)O)O)C(C(C(O1)C)O)O |
M END | M END | ||
| − | |||
Latest revision as of 09:00, 14 March 2009
Copyright: ARM project http://www.metabolome.jp/
44 48 0 0 0 0 0 0 0 0999 V2000
-1.6648 0.1873 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.6648 -0.6381 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.9500 -1.0506 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.2351 -0.6381 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.2352 0.1874 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.9500 0.6001 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.4797 -1.0507 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.1944 -0.6381 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.1944 0.1874 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.4797 0.6001 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.4798 -1.6942 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.0946 0.7589 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.8231 0.3383 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.5515 0.7590 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.5516 1.6002 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.8231 2.0208 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.0946 1.6001 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.9499 -1.8758 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
4.2326 2.0763 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.5186 0.6804 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.0344 -1.1686 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.1385 -1.4595 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.4504 -1.7804 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.1680 -2.0281 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.6498 -2.6202 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.3380 -2.2994 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.6204 -2.0516 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.5245 -1.0656 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
4.4413 -1.9019 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
4.7584 -2.5862 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.9534 0.4528 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.1858 0.3173 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.6434 0.9483 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.6999 1.7299 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.4674 1.8653 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.0100 1.2343 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.4582 2.4822 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.8603 2.2041 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.7584 1.3431 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.5384 0.2573 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
4.2403 -2.9107 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.6882 -3.8715 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.8174 2.7185 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.3846 3.8715 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1 2 1 0 0 0 0
2 3 2 0 0 0 0
4 5 2 0 0 0 0
5 6 1 0 0 0 0
6 1 2 0 0 0 0
4 7 1 0 0 0 0
7 8 1 0 0 0 0
8 9 2 0 0 0 0
9 10 1 0 0 0 0
10 5 1 0 0 0 0
7 11 2 0 0 0 0
9 12 1 0 0 0 0
12 13 2 0 0 0 0
13 14 1 0 0 0 0
14 15 2 0 0 0 0
15 16 1 0 0 0 0
16 17 2 0 0 0 0
17 12 1 0 0 0 0
3 18 1 0 0 0 0
19 15 1 0 0 0 0
4 3 1 0 0 0 0
1 20 1 0 0 0 0
21 8 1 0 0 0 0
22 23 1 0 0 0 0
23 24 1 1 0 0 0
24 25 1 1 0 0 0
26 25 1 1 0 0 0
26 27 1 0 0 0 0
27 22 1 0 0 0 0
22 28 1 0 0 0 0
27 29 1 0 0 0 0
26 30 1 0 0 0 0
21 23 1 0 0 0 0
32 31 1 1 0 0 0
32 33 1 0 0 0 0
33 34 1 0 0 0 0
34 35 1 0 0 0 0
35 36 1 1 0 0 0
36 31 1 1 0 0 0
35 37 1 0 0 0 0
34 38 1 0 0 0 0
36 39 1 0 0 0 0
31 40 1 0 0 0 0
32 20 1 0 0 0 0
41 42 1 0 0 0 0
25 41 1 0 0 0 0
43 44 1 0 0 0 0
16 43 1 0 0 0 0
M STY 1 1 SUP
M SLB 1 1 1
M SAL 1 2 41 42
M SBL 1 1 46
M SMT 1 CH2OH
M SBV 1 46 -0.5904 0.2905
M STY 1 2 SUP
M SLB 1 2 2
M SAL 2 2 43 44
M SBL 2 1 48
M SMT 2 OCH3
M SBV 2 48 0.0057 -0.6976
S SKP 5
ID FL5FADGL0015
FORMULA C28H32O16
EXACTMASS 624.1690349759999
AVERAGEMASS 624.54408
SMILES OC(C1Oc(c5)cc(c3c5O)OC(c(c4)cc(OC)c(O)c4)=C(C3=O)OC(O2)C(C(C(O)C2CO)O)O)C(C(C(O1)C)O)O
M END