Mol:FL5FADGA0022
From Metabolomics.JP
(Difference between revisions)
| Line 1: | Line 1: | ||
| − | + | ||
| − | + | ||
| − | Copyright: ARM project http://www.metabolome.jp/ | + | Copyright: ARM project http://www.metabolome.jp/ |
| − | 51 55 0 0 0 0 0 0 0 0999 V2000 | + | 51 55 0 0 0 0 0 0 0 0999 V2000 |
| − | -3.0855 1.7742 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.0855 1.7742 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.0855 1.1319 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.0855 1.1319 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.5292 0.8107 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.5292 0.8107 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.9729 1.1319 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.9729 1.1319 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.9729 1.7742 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.9729 1.7742 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.5292 2.0954 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.5292 2.0954 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.4166 0.8107 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.4166 0.8107 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.8603 1.1319 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.8603 1.1319 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.8603 1.7742 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.8603 1.7742 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.4166 2.0954 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.4166 2.0954 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.4166 0.3099 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.4166 0.3099 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.1598 2.2191 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.1598 2.2191 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.4072 1.8917 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.4072 1.8917 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.9742 2.2191 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.9742 2.2191 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.9742 2.8737 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.9742 2.8737 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.4072 3.2011 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.4072 3.2011 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.1598 2.8737 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.1598 2.8737 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.5292 0.1686 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.5292 0.1686 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.8403 3.3736 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.8403 3.3736 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.6011 2.2280 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.6011 2.2280 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.5106 1.3265 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0 | + | 0.5106 1.3265 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0 |
| − | 0.9123 0.7963 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | 0.9123 0.7963 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | 1.4907 1.0212 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | 1.4907 1.0212 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | 2.0489 1.0273 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | 2.0489 1.0273 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | 1.7464 1.4536 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 | + | 1.7464 1.4536 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 |
| − | 1.1372 1.1658 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.1372 1.1658 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.0455 1.0055 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.0455 1.0055 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.5331 0.2971 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.5331 0.2971 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.4431 0.3753 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.4431 0.3753 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.4592 0.8360 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.4592 0.8360 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.5145 -0.8373 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 | + | 1.5145 -0.8373 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 |
| − | 1.2445 -0.2583 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | 1.2445 -0.2583 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | 1.0361 -0.8621 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.0361 -0.8621 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.5377 -1.2676 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | 0.5377 -1.2676 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | 0.8077 -1.8467 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | 0.8077 -1.8467 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | 1.0163 -1.2428 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 | + | 1.0163 -1.2428 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 |
| − | 1.7246 -0.2601 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.7246 -0.2601 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.4038 -1.5680 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.4038 -1.5680 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.4990 -2.1057 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.4990 -2.1057 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.1023 -2.1816 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.1023 -2.1816 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.8200 -2.5164 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.8200 -2.5164 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.3700 -2.1312 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.3700 -2.1312 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.3277 -1.6489 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.3277 -1.6489 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.8261 -2.5140 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.8261 -2.5140 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.0509 -2.4921 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.0509 -2.4921 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.6853 -2.3803 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.6853 -2.3803 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.8408 -3.0694 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.8408 -3.0694 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.6906 3.7772 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.6906 3.7772 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.1320 4.6745 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.1320 4.6745 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.9771 1.6956 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.9771 1.6956 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.7524 1.9778 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.7524 1.9778 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1 2 1 0 0 0 0 | + | 1 2 1 0 0 0 0 |
| − | 2 3 2 0 0 0 0 | + | 2 3 2 0 0 0 0 |
| − | 4 5 2 0 0 0 0 | + | 4 5 2 0 0 0 0 |
| − | 5 6 1 0 0 0 0 | + | 5 6 1 0 0 0 0 |
| − | 6 1 2 0 0 0 0 | + | 6 1 2 0 0 0 0 |
| − | 4 7 1 0 0 0 0 | + | 4 7 1 0 0 0 0 |
| − | 7 8 1 0 0 0 0 | + | 7 8 1 0 0 0 0 |
| − | 8 9 2 0 0 0 0 | + | 8 9 2 0 0 0 0 |
| − | 9 10 1 0 0 0 0 | + | 9 10 1 0 0 0 0 |
| − | 10 5 1 0 0 0 0 | + | 10 5 1 0 0 0 0 |
| − | 7 11 2 0 0 0 0 | + | 7 11 2 0 0 0 0 |
| − | 9 12 1 0 0 0 0 | + | 9 12 1 0 0 0 0 |
| − | 12 13 2 0 0 0 0 | + | 12 13 2 0 0 0 0 |
| − | 13 14 1 0 0 0 0 | + | 13 14 1 0 0 0 0 |
| − | 14 15 2 0 0 0 0 | + | 14 15 2 0 0 0 0 |
| − | 15 16 1 0 0 0 0 | + | 15 16 1 0 0 0 0 |
| − | 16 17 2 0 0 0 0 | + | 16 17 2 0 0 0 0 |
| − | 17 12 1 0 0 0 0 | + | 17 12 1 0 0 0 0 |
| − | 3 18 1 0 0 0 0 | + | 3 18 1 0 0 0 0 |
| − | 19 15 1 0 0 0 0 | + | 19 15 1 0 0 0 0 |
| − | 4 3 1 0 0 0 0 | + | 4 3 1 0 0 0 0 |
| − | 1 20 1 0 0 0 0 | + | 1 20 1 0 0 0 0 |
| − | 21 22 1 1 0 0 0 | + | 21 22 1 1 0 0 0 |
| − | 22 23 1 1 0 0 0 | + | 22 23 1 1 0 0 0 |
| − | 24 23 1 1 0 0 0 | + | 24 23 1 1 0 0 0 |
| − | 24 25 1 0 0 0 0 | + | 24 25 1 0 0 0 0 |
| − | 25 26 1 0 0 0 0 | + | 25 26 1 0 0 0 0 |
| − | 26 21 1 0 0 0 0 | + | 26 21 1 0 0 0 0 |
| − | 21 27 1 0 0 0 0 | + | 21 27 1 0 0 0 0 |
| − | 22 28 1 0 0 0 0 | + | 22 28 1 0 0 0 0 |
| − | 23 29 1 0 0 0 0 | + | 23 29 1 0 0 0 0 |
| − | 24 30 1 0 0 0 0 | + | 24 30 1 0 0 0 0 |
| − | 27 8 1 0 0 0 0 | + | 27 8 1 0 0 0 0 |
| − | 31 32 1 0 0 0 0 | + | 31 32 1 0 0 0 0 |
| − | 32 33 1 1 0 0 0 | + | 32 33 1 1 0 0 0 |
| − | 33 34 1 1 0 0 0 | + | 33 34 1 1 0 0 0 |
| − | 35 34 1 1 0 0 0 | + | 35 34 1 1 0 0 0 |
| − | 35 36 1 0 0 0 0 | + | 35 36 1 0 0 0 0 |
| − | 36 31 1 0 0 0 0 | + | 36 31 1 0 0 0 0 |
| − | 31 37 1 0 0 0 0 | + | 31 37 1 0 0 0 0 |
| − | 36 38 1 0 0 0 0 | + | 36 38 1 0 0 0 0 |
| − | 35 39 1 0 0 0 0 | + | 35 39 1 0 0 0 0 |
| − | 34 40 1 0 0 0 0 | + | 34 40 1 0 0 0 0 |
| − | 32 29 1 0 0 0 0 | + | 32 29 1 0 0 0 0 |
| − | 40 41 1 0 0 0 0 | + | 40 41 1 0 0 0 0 |
| − | 41 42 1 0 0 0 0 | + | 41 42 1 0 0 0 0 |
| − | 42 43 2 0 0 0 0 | + | 42 43 2 0 0 0 0 |
| − | 42 44 1 0 0 0 0 | + | 42 44 1 0 0 0 0 |
| − | 39 45 1 0 0 0 0 | + | 39 45 1 0 0 0 0 |
| − | 45 46 2 0 0 0 0 | + | 45 46 2 0 0 0 0 |
| − | 45 47 1 0 0 0 0 | + | 45 47 1 0 0 0 0 |
| − | 16 48 1 0 0 0 0 | + | 16 48 1 0 0 0 0 |
| − | 48 49 1 0 0 0 0 | + | 48 49 1 0 0 0 0 |
| − | 25 50 1 0 0 0 0 | + | 25 50 1 0 0 0 0 |
| − | 50 51 1 0 0 0 0 | + | 50 51 1 0 0 0 0 |
| − | M STY 1 2 SUP | + | M STY 1 2 SUP |
| − | M SLB 1 2 2 | + | M SLB 1 2 2 |
| − | M SAL 2 2 50 51 | + | M SAL 2 2 50 51 |
| − | M SBL 2 1 54 | + | M SBL 2 1 54 |
| − | M SMT 2 CH2OH | + | M SMT 2 CH2OH |
| − | M SVB 2 54 2.0801 1.4324 | + | M SVB 2 54 2.0801 1.4324 |
| − | M STY 1 1 SUP | + | M STY 1 1 SUP |
| − | M SLB 1 1 1 | + | M SLB 1 1 1 |
| − | M SAL 1 2 48 49 | + | M SAL 1 2 48 49 |
| − | M SBL 1 1 52 | + | M SBL 1 1 52 |
| − | M SMT 1 OCH3 | + | M SMT 1 OCH3 |
| − | M SVB 1 52 0.6906 3.7772 | + | M SVB 1 52 0.6906 3.7772 |
| − | S SKP 8 | + | S SKP 8 |
| − | ID FL5FADGA0022 | + | ID FL5FADGA0022 |
| − | KNApSAcK_ID C00006013 | + | KNApSAcK_ID C00006013 |
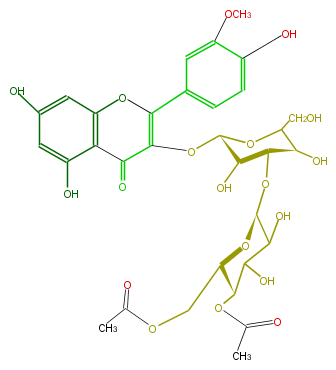
| − | NAME Isorhamnetin 3-(4''',6'''-diacetylglucosyl) (1->3)-galactoside | + | NAME Isorhamnetin 3-(4''',6'''-diacetylglucosyl) (1->3)-galactoside |
| − | CAS_RN 139955-73-0 | + | CAS_RN 139955-73-0 |
| − | FORMULA C32H36O19 | + | FORMULA C32H36O19 |
| − | EXACTMASS 724.18507897 | + | EXACTMASS 724.18507897 |
| − | AVERAGEMASS 724.61684 | + | AVERAGEMASS 724.61684 |
| − | SMILES O(C(c(c5)ccc(c(OC)5)O)=1)c(c4)c(c(O)cc4O)C(=O)C1O[C@H]([C@@H]2O)OC([C@@H](O)[C@H]2O[C@@H](C3O)O[C@H](COC(C)=O)[C@H](OC(C)=O)C3O)CO | + | SMILES O(C(c(c5)ccc(c(OC)5)O)=1)c(c4)c(c(O)cc4O)C(=O)C1O[C@H]([C@@H]2O)OC([C@@H](O)[C@H]2O[C@@H](C3O)O[C@H](COC(C)=O)[C@H](OC(C)=O)C3O)CO |
M END | M END | ||
| − | |||
Latest revision as of 09:00, 14 March 2009
Copyright: ARM project http://www.metabolome.jp/
51 55 0 0 0 0 0 0 0 0999 V2000
-3.0855 1.7742 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.0855 1.1319 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.5292 0.8107 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.9729 1.1319 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.9729 1.7742 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.5292 2.0954 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.4166 0.8107 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.8603 1.1319 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.8603 1.7742 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.4166 2.0954 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.4166 0.3099 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.1598 2.2191 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.4072 1.8917 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.9742 2.2191 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.9742 2.8737 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.4072 3.2011 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.1598 2.8737 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.5292 0.1686 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.8403 3.3736 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.6011 2.2280 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.5106 1.3265 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0
0.9123 0.7963 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
1.4907 1.0212 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
2.0489 1.0273 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
1.7464 1.4536 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0
1.1372 1.1658 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.0455 1.0055 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.5331 0.2971 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.4431 0.3753 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.4592 0.8360 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.5145 -0.8373 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0
1.2445 -0.2583 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
1.0361 -0.8621 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.5377 -1.2676 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
0.8077 -1.8467 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
1.0163 -1.2428 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0
1.7246 -0.2601 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.4038 -1.5680 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.4990 -2.1057 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.1023 -2.1816 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.8200 -2.5164 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.3700 -2.1312 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.3277 -1.6489 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.8261 -2.5140 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.0509 -2.4921 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.6853 -2.3803 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.8408 -3.0694 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.6906 3.7772 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.1320 4.6745 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.9771 1.6956 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.7524 1.9778 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1 2 1 0 0 0 0
2 3 2 0 0 0 0
4 5 2 0 0 0 0
5 6 1 0 0 0 0
6 1 2 0 0 0 0
4 7 1 0 0 0 0
7 8 1 0 0 0 0
8 9 2 0 0 0 0
9 10 1 0 0 0 0
10 5 1 0 0 0 0
7 11 2 0 0 0 0
9 12 1 0 0 0 0
12 13 2 0 0 0 0
13 14 1 0 0 0 0
14 15 2 0 0 0 0
15 16 1 0 0 0 0
16 17 2 0 0 0 0
17 12 1 0 0 0 0
3 18 1 0 0 0 0
19 15 1 0 0 0 0
4 3 1 0 0 0 0
1 20 1 0 0 0 0
21 22 1 1 0 0 0
22 23 1 1 0 0 0
24 23 1 1 0 0 0
24 25 1 0 0 0 0
25 26 1 0 0 0 0
26 21 1 0 0 0 0
21 27 1 0 0 0 0
22 28 1 0 0 0 0
23 29 1 0 0 0 0
24 30 1 0 0 0 0
27 8 1 0 0 0 0
31 32 1 0 0 0 0
32 33 1 1 0 0 0
33 34 1 1 0 0 0
35 34 1 1 0 0 0
35 36 1 0 0 0 0
36 31 1 0 0 0 0
31 37 1 0 0 0 0
36 38 1 0 0 0 0
35 39 1 0 0 0 0
34 40 1 0 0 0 0
32 29 1 0 0 0 0
40 41 1 0 0 0 0
41 42 1 0 0 0 0
42 43 2 0 0 0 0
42 44 1 0 0 0 0
39 45 1 0 0 0 0
45 46 2 0 0 0 0
45 47 1 0 0 0 0
16 48 1 0 0 0 0
48 49 1 0 0 0 0
25 50 1 0 0 0 0
50 51 1 0 0 0 0
M STY 1 2 SUP
M SLB 1 2 2
M SAL 2 2 50 51
M SBL 2 1 54
M SMT 2 CH2OH
M SVB 2 54 2.0801 1.4324
M STY 1 1 SUP
M SLB 1 1 1
M SAL 1 2 48 49
M SBL 1 1 52
M SMT 1 OCH3
M SVB 1 52 0.6906 3.7772
S SKP 8
ID FL5FADGA0022
KNApSAcK_ID C00006013
NAME Isorhamnetin 3-(4''',6'''-diacetylglucosyl) (1->3)-galactoside
CAS_RN 139955-73-0
FORMULA C32H36O19
EXACTMASS 724.18507897
AVERAGEMASS 724.61684
SMILES O(C(c(c5)ccc(c(OC)5)O)=1)c(c4)c(c(O)cc4O)C(=O)C1O[C@H]([C@@H]2O)OC([C@@H](O)[C@H]2O[C@@H](C3O)O[C@H](COC(C)=O)[C@H](OC(C)=O)C3O)CO
M END