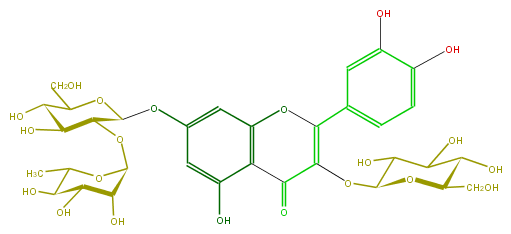
Mol:FL5FACGL0047
From Metabolomics.JP
(Difference between revisions)
| Line 1: | Line 1: | ||
| − | + | ||
| − | + | ||
| − | Copyright: ARM project http://www.metabolome.jp/ | + | Copyright: ARM project http://www.metabolome.jp/ |
| − | 54 59 0 0 0 0 0 0 0 0999 V2000 | + | 54 59 0 0 0 0 0 0 0 0999 V2000 |
| − | -1.2916 -0.0818 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.2916 -0.0818 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.2916 -0.9627 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.2916 -0.9627 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.5288 -1.4031 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.5288 -1.4031 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.2339 -0.9627 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.2339 -0.9627 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.2339 -0.0818 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.2339 -0.0818 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.5288 0.3586 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.5288 0.3586 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.9967 -1.4031 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.9967 -1.4031 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.7595 -0.9627 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.7595 -0.9627 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.7595 -0.0818 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.7595 -0.0818 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.9967 0.3586 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.9967 0.3586 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.9967 -2.0898 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.9967 -2.0898 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.4931 0.3837 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.4931 0.3837 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.2705 -0.0651 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.2705 -0.0651 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.0480 0.3837 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.0480 0.3837 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.0480 1.2814 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.0480 1.2814 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.2705 1.7302 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.2705 1.7302 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.4931 1.2814 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.4931 1.2814 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.5288 -2.2835 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.5288 -2.2835 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.8800 1.7617 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.8800 1.7617 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.5167 -1.4059 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.5167 -1.4059 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.2705 2.6276 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.2705 2.6276 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.5965 -0.8142 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.5965 -0.8142 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.1839 -1.5285 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.1839 -1.5285 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.9771 -1.3018 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.9771 -1.3018 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.7751 -1.5285 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.7751 -1.5285 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 5.1877 -0.8142 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 5.1877 -0.8142 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.3945 -1.0407 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.3945 -1.0407 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.9776 -0.8984 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.9776 -0.8984 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.9684 -0.3914 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.9684 -0.3914 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 5.9079 -1.0560 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 5.9079 -1.0560 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.0803 0.3735 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.0803 0.3735 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.6864 0.4477 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.6864 0.4477 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.2116 -0.1789 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.2116 -0.1789 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.5279 0.0870 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.5279 0.0870 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.8154 0.0790 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.8154 0.0790 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.3476 0.5734 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.3476 0.5734 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.0459 0.3228 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.0459 0.3228 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -5.2349 0.1945 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -5.2349 0.1945 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.9916 -0.1339 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.9916 -0.1339 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.8874 -0.3658 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.8874 -0.3658 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.0786 -1.0727 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.0786 -1.0727 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.4355 -1.6910 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.4355 -1.6910 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.7493 -1.4947 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.7493 -1.4947 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.0586 -1.6910 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.0586 -1.6910 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.7017 -1.0727 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.7017 -1.0727 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.3881 -1.2688 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.3881 -1.2688 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.7655 -1.1624 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.7655 -1.1624 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.9355 -1.5854 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.9355 -1.5854 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.2999 -2.0875 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.2999 -2.0875 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.0291 -2.3062 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.0291 -2.3062 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 5.3816 -1.4962 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 5.3816 -1.4962 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 5.3816 -2.6276 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 5.3816 -2.6276 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.5807 0.9260 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.5807 0.9260 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -5.9079 1.2699 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -5.9079 1.2699 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1 2 1 0 0 0 0 | + | 1 2 1 0 0 0 0 |
| − | 2 3 2 0 0 0 0 | + | 2 3 2 0 0 0 0 |
| − | 4 5 2 0 0 0 0 | + | 4 5 2 0 0 0 0 |
| − | 5 6 1 0 0 0 0 | + | 5 6 1 0 0 0 0 |
| − | 6 1 2 0 0 0 0 | + | 6 1 2 0 0 0 0 |
| − | 4 7 1 0 0 0 0 | + | 4 7 1 0 0 0 0 |
| − | 7 8 1 0 0 0 0 | + | 7 8 1 0 0 0 0 |
| − | 8 9 2 0 0 0 0 | + | 8 9 2 0 0 0 0 |
| − | 9 10 1 0 0 0 0 | + | 9 10 1 0 0 0 0 |
| − | 10 5 1 0 0 0 0 | + | 10 5 1 0 0 0 0 |
| − | 7 11 2 0 0 0 0 | + | 7 11 2 0 0 0 0 |
| − | 9 12 1 0 0 0 0 | + | 9 12 1 0 0 0 0 |
| − | 12 13 2 0 0 0 0 | + | 12 13 2 0 0 0 0 |
| − | 13 14 1 0 0 0 0 | + | 13 14 1 0 0 0 0 |
| − | 14 15 2 0 0 0 0 | + | 14 15 2 0 0 0 0 |
| − | 15 16 1 0 0 0 0 | + | 15 16 1 0 0 0 0 |
| − | 16 17 2 0 0 0 0 | + | 16 17 2 0 0 0 0 |
| − | 17 12 1 0 0 0 0 | + | 17 12 1 0 0 0 0 |
| − | 3 18 1 0 0 0 0 | + | 3 18 1 0 0 0 0 |
| − | 19 15 1 0 0 0 0 | + | 19 15 1 0 0 0 0 |
| − | 20 8 1 0 0 0 0 | + | 20 8 1 0 0 0 0 |
| − | 4 3 1 0 0 0 0 | + | 4 3 1 0 0 0 0 |
| − | 16 21 1 0 0 0 0 | + | 16 21 1 0 0 0 0 |
| − | 22 23 1 0 0 0 0 | + | 22 23 1 0 0 0 0 |
| − | 23 24 1 1 0 0 0 | + | 23 24 1 1 0 0 0 |
| − | 24 25 1 1 0 0 0 | + | 24 25 1 1 0 0 0 |
| − | 26 25 1 1 0 0 0 | + | 26 25 1 1 0 0 0 |
| − | 26 27 1 0 0 0 0 | + | 26 27 1 0 0 0 0 |
| − | 27 22 1 0 0 0 0 | + | 27 22 1 0 0 0 0 |
| − | 22 28 1 0 0 0 0 | + | 22 28 1 0 0 0 0 |
| − | 27 29 1 0 0 0 0 | + | 27 29 1 0 0 0 0 |
| − | 26 30 1 0 0 0 0 | + | 26 30 1 0 0 0 0 |
| − | 23 20 1 0 0 0 0 | + | 23 20 1 0 0 0 0 |
| − | 1 31 1 0 0 0 0 | + | 1 31 1 0 0 0 0 |
| − | 32 33 1 1 0 0 0 | + | 32 33 1 1 0 0 0 |
| − | 33 34 1 1 0 0 0 | + | 33 34 1 1 0 0 0 |
| − | 35 34 1 1 0 0 0 | + | 35 34 1 1 0 0 0 |
| − | 35 36 1 0 0 0 0 | + | 35 36 1 0 0 0 0 |
| − | 36 37 1 0 0 0 0 | + | 36 37 1 0 0 0 0 |
| − | 37 32 1 0 0 0 0 | + | 37 32 1 0 0 0 0 |
| − | 32 38 1 0 0 0 0 | + | 32 38 1 0 0 0 0 |
| − | 33 39 1 0 0 0 0 | + | 33 39 1 0 0 0 0 |
| − | 34 40 1 0 0 0 0 | + | 34 40 1 0 0 0 0 |
| − | 41 42 1 0 0 0 0 | + | 41 42 1 0 0 0 0 |
| − | 42 43 1 1 0 0 0 | + | 42 43 1 1 0 0 0 |
| − | 43 44 1 1 0 0 0 | + | 43 44 1 1 0 0 0 |
| − | 45 44 1 1 0 0 0 | + | 45 44 1 1 0 0 0 |
| − | 45 46 1 0 0 0 0 | + | 45 46 1 0 0 0 0 |
| − | 46 41 1 0 0 0 0 | + | 46 41 1 0 0 0 0 |
| − | 41 47 1 0 0 0 0 | + | 41 47 1 0 0 0 0 |
| − | 42 48 1 0 0 0 0 | + | 42 48 1 0 0 0 0 |
| − | 43 49 1 0 0 0 0 | + | 43 49 1 0 0 0 0 |
| − | 44 50 1 0 0 0 0 | + | 44 50 1 0 0 0 0 |
| − | 45 40 1 0 0 0 0 | + | 45 40 1 0 0 0 0 |
| − | 35 31 1 0 0 0 0 | + | 35 31 1 0 0 0 0 |
| − | 51 52 1 0 0 0 0 | + | 51 52 1 0 0 0 0 |
| − | 25 51 1 0 0 0 0 | + | 25 51 1 0 0 0 0 |
| − | 53 54 1 0 0 0 0 | + | 53 54 1 0 0 0 0 |
| − | 37 53 1 0 0 0 0 | + | 37 53 1 0 0 0 0 |
| − | M STY 1 1 SUP | + | M STY 1 1 SUP |
| − | M SLB 1 1 1 | + | M SLB 1 1 1 |
| − | M SAL 1 2 51 52 | + | M SAL 1 2 51 52 |
| − | M SBL 1 1 57 | + | M SBL 1 1 57 |
| − | M SMT 1 CH2OH | + | M SMT 1 CH2OH |
| − | M SBV 1 57 -0.6064 -0.0323 | + | M SBV 1 57 -0.6064 -0.0323 |
| − | M STY 1 2 SUP | + | M STY 1 2 SUP |
| − | M SLB 1 2 2 | + | M SLB 1 2 2 |
| − | M SAL 2 2 53 54 | + | M SAL 2 2 53 54 |
| − | M SBL 2 1 59 | + | M SBL 2 1 59 |
| − | M SMT 2 ^ CH2OH | + | M SMT 2 ^ CH2OH |
| − | M SBV 2 59 0.5347 -0.6032 | + | M SBV 2 59 0.5347 -0.6032 |
| − | S SKP 5 | + | S SKP 5 |
| − | ID FL5FACGL0047 | + | ID FL5FACGL0047 |
| − | FORMULA C33H40O21 | + | FORMULA C33H40O21 |
| − | EXACTMASS 772.206208342 | + | EXACTMASS 772.206208342 |
| − | AVERAGEMASS 772.6581 | + | AVERAGEMASS 772.6581 |
| − | SMILES OC(C(O)5)C(CO)OC(C5OC(O6)C(C(C(O)C(C)6)O)O)Oc(c4)cc(c(c4O)1)OC(c(c3)ccc(c3O)O)=C(OC(O2)C(O)C(C(O)C2CO)O)C1=O | + | SMILES OC(C(O)5)C(CO)OC(C5OC(O6)C(C(C(O)C(C)6)O)O)Oc(c4)cc(c(c4O)1)OC(c(c3)ccc(c3O)O)=C(OC(O2)C(O)C(C(O)C2CO)O)C1=O |
M END | M END | ||
| − | |||
Latest revision as of 09:00, 14 March 2009
Copyright: ARM project http://www.metabolome.jp/
54 59 0 0 0 0 0 0 0 0999 V2000
-1.2916 -0.0818 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.2916 -0.9627 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.5288 -1.4031 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.2339 -0.9627 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.2339 -0.0818 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.5288 0.3586 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.9967 -1.4031 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.7595 -0.9627 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.7595 -0.0818 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.9967 0.3586 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.9967 -2.0898 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.4931 0.3837 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.2705 -0.0651 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.0480 0.3837 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.0480 1.2814 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.2705 1.7302 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.4931 1.2814 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.5288 -2.2835 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
4.8800 1.7617 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.5167 -1.4059 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.2705 2.6276 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.5965 -0.8142 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.1839 -1.5285 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.9771 -1.3018 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
4.7751 -1.5285 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
5.1877 -0.8142 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.3945 -1.0407 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.9776 -0.8984 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
4.9684 -0.3914 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
5.9079 -1.0560 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.0803 0.3735 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.6864 0.4477 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.2116 -0.1789 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.5279 0.0870 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.8154 0.0790 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.3476 0.5734 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.0459 0.3228 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-5.2349 0.1945 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.9916 -0.1339 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.8874 -0.3658 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.0786 -1.0727 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.4355 -1.6910 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.7493 -1.4947 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.0586 -1.6910 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.7017 -1.0727 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.3881 -1.2688 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.7655 -1.1624 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.9355 -1.5854 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.2999 -2.0875 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.0291 -2.3062 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
5.3816 -1.4962 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
5.3816 -2.6276 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.5807 0.9260 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-5.9079 1.2699 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1 2 1 0 0 0 0
2 3 2 0 0 0 0
4 5 2 0 0 0 0
5 6 1 0 0 0 0
6 1 2 0 0 0 0
4 7 1 0 0 0 0
7 8 1 0 0 0 0
8 9 2 0 0 0 0
9 10 1 0 0 0 0
10 5 1 0 0 0 0
7 11 2 0 0 0 0
9 12 1 0 0 0 0
12 13 2 0 0 0 0
13 14 1 0 0 0 0
14 15 2 0 0 0 0
15 16 1 0 0 0 0
16 17 2 0 0 0 0
17 12 1 0 0 0 0
3 18 1 0 0 0 0
19 15 1 0 0 0 0
20 8 1 0 0 0 0
4 3 1 0 0 0 0
16 21 1 0 0 0 0
22 23 1 0 0 0 0
23 24 1 1 0 0 0
24 25 1 1 0 0 0
26 25 1 1 0 0 0
26 27 1 0 0 0 0
27 22 1 0 0 0 0
22 28 1 0 0 0 0
27 29 1 0 0 0 0
26 30 1 0 0 0 0
23 20 1 0 0 0 0
1 31 1 0 0 0 0
32 33 1 1 0 0 0
33 34 1 1 0 0 0
35 34 1 1 0 0 0
35 36 1 0 0 0 0
36 37 1 0 0 0 0
37 32 1 0 0 0 0
32 38 1 0 0 0 0
33 39 1 0 0 0 0
34 40 1 0 0 0 0
41 42 1 0 0 0 0
42 43 1 1 0 0 0
43 44 1 1 0 0 0
45 44 1 1 0 0 0
45 46 1 0 0 0 0
46 41 1 0 0 0 0
41 47 1 0 0 0 0
42 48 1 0 0 0 0
43 49 1 0 0 0 0
44 50 1 0 0 0 0
45 40 1 0 0 0 0
35 31 1 0 0 0 0
51 52 1 0 0 0 0
25 51 1 0 0 0 0
53 54 1 0 0 0 0
37 53 1 0 0 0 0
M STY 1 1 SUP
M SLB 1 1 1
M SAL 1 2 51 52
M SBL 1 1 57
M SMT 1 CH2OH
M SBV 1 57 -0.6064 -0.0323
M STY 1 2 SUP
M SLB 1 2 2
M SAL 2 2 53 54
M SBL 2 1 59
M SMT 2 ^ CH2OH
M SBV 2 59 0.5347 -0.6032
S SKP 5
ID FL5FACGL0047
FORMULA C33H40O21
EXACTMASS 772.206208342
AVERAGEMASS 772.6581
SMILES OC(C(O)5)C(CO)OC(C5OC(O6)C(C(C(O)C(C)6)O)O)Oc(c4)cc(c(c4O)1)OC(c(c3)ccc(c3O)O)=C(OC(O2)C(O)C(C(O)C2CO)O)C1=O
M END