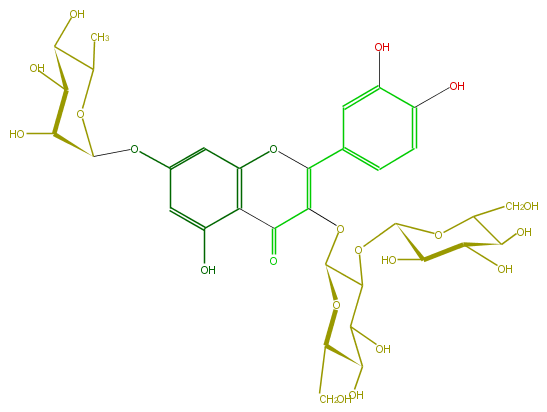
Mol:FL5FACGL0034
From Metabolomics.JP
(Difference between revisions)
| Line 1: | Line 1: | ||
| − | + | ||
| − | + | ||
| − | Copyright: ARM project http://www.metabolome.jp/ | + | Copyright: ARM project http://www.metabolome.jp/ |
| − | 54 59 0 0 0 0 0 0 0 0999 V2000 | + | 54 59 0 0 0 0 0 0 0 0999 V2000 |
| − | -2.0380 0.7467 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.0380 0.7467 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.0380 -0.0629 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.0380 -0.0629 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.3368 -0.4677 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.3368 -0.4677 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.6357 -0.0629 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.6357 -0.0629 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.6357 0.7467 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.6357 0.7467 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.3368 1.1515 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.3368 1.1515 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.0654 -0.4677 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.0654 -0.4677 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.7665 -0.0629 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.7665 -0.0629 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.7665 0.7467 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.7665 0.7467 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.0654 1.1515 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.0654 1.1515 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.0654 -1.0989 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.0654 -1.0989 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.4674 1.1513 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.4674 1.1513 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.1818 0.7388 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.1818 0.7388 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.8964 1.1513 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.8964 1.1513 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.8964 1.9764 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.8964 1.9764 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.1818 2.3890 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.1818 2.3890 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.4674 1.9764 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.4674 1.9764 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.7388 1.1513 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.7388 1.1513 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.4103 -0.4527 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.4103 -0.4527 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.3368 -1.2770 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.3368 -1.2770 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.6953 2.4377 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.6953 2.4377 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.8380 -1.6111 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.8380 -1.6111 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.1034 -1.1871 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.1034 -1.1871 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.3366 -2.0025 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.3366 -2.0025 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.1034 -2.8230 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.1034 -2.8230 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.8380 -3.2471 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.8380 -3.2471 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.6051 -2.4315 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.6051 -2.4315 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.7727 -0.8868 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.7727 -0.8868 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.2086 -2.8719 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.2086 -2.8719 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.6644 -3.8168 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.6644 -3.8168 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.5227 -0.3619 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.5227 -0.3619 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.0803 -1.0979 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.0803 -1.0979 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.8833 -0.7856 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.8833 -0.7856 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.6580 -0.7773 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.6580 -0.7773 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.0950 -0.2142 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.0950 -0.2142 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.3926 -0.5848 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.3926 -0.5848 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.4601 -1.0796 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.4601 -1.0796 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.6540 -1.2706 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.6540 -1.2706 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 5.0475 -0.5116 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 5.0475 -0.5116 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.1818 3.2139 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.1818 3.2139 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.3800 1.4462 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.3800 1.4462 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.5882 0.9891 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.5882 0.9891 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.8395 1.8682 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.8395 1.8682 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.5882 2.7525 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.5882 2.7525 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.3800 3.2098 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.3800 3.2098 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.1289 2.3307 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.1289 2.3307 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.9727 3.8890 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.9727 3.8890 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.5689 3.4247 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.5689 3.4247 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.7778 2.7883 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.7778 2.7883 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -5.0475 1.4513 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -5.0475 1.4513 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.8234 0.0099 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.8234 0.0099 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.5871 1.2479 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.5871 1.2479 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.9615 -3.8890 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.9615 -3.8890 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.2911 -3.7502 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.2911 -3.7502 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1 2 1 0 0 0 0 | + | 1 2 1 0 0 0 0 |
| − | 2 3 2 0 0 0 0 | + | 2 3 2 0 0 0 0 |
| − | 3 4 1 0 0 0 0 | + | 3 4 1 0 0 0 0 |
| − | 4 5 2 0 0 0 0 | + | 4 5 2 0 0 0 0 |
| − | 5 6 1 0 0 0 0 | + | 5 6 1 0 0 0 0 |
| − | 6 1 2 0 0 0 0 | + | 6 1 2 0 0 0 0 |
| − | 4 7 1 0 0 0 0 | + | 4 7 1 0 0 0 0 |
| − | 7 8 1 0 0 0 0 | + | 7 8 1 0 0 0 0 |
| − | 8 9 2 0 0 0 0 | + | 8 9 2 0 0 0 0 |
| − | 9 10 1 0 0 0 0 | + | 9 10 1 0 0 0 0 |
| − | 10 5 1 0 0 0 0 | + | 10 5 1 0 0 0 0 |
| − | 7 11 2 0 0 0 0 | + | 7 11 2 0 0 0 0 |
| − | 9 12 1 0 0 0 0 | + | 9 12 1 0 0 0 0 |
| − | 12 13 2 0 0 0 0 | + | 12 13 2 0 0 0 0 |
| − | 13 14 1 0 0 0 0 | + | 13 14 1 0 0 0 0 |
| − | 14 15 2 0 0 0 0 | + | 14 15 2 0 0 0 0 |
| − | 15 16 1 0 0 0 0 | + | 15 16 1 0 0 0 0 |
| − | 16 17 2 0 0 0 0 | + | 16 17 2 0 0 0 0 |
| − | 17 12 1 0 0 0 0 | + | 17 12 1 0 0 0 0 |
| − | 1 18 1 0 0 0 0 | + | 1 18 1 0 0 0 0 |
| − | 8 19 1 0 0 0 0 | + | 8 19 1 0 0 0 0 |
| − | 3 20 1 0 0 0 0 | + | 3 20 1 0 0 0 0 |
| − | 21 15 1 0 0 0 0 | + | 21 15 1 0 0 0 0 |
| − | 22 23 1 0 0 0 0 | + | 22 23 1 0 0 0 0 |
| − | 23 24 1 1 0 0 0 | + | 23 24 1 1 0 0 0 |
| − | 24 25 1 1 0 0 0 | + | 24 25 1 1 0 0 0 |
| − | 26 25 1 1 0 0 0 | + | 26 25 1 1 0 0 0 |
| − | 26 27 1 0 0 0 0 | + | 26 27 1 0 0 0 0 |
| − | 27 22 1 0 0 0 0 | + | 27 22 1 0 0 0 0 |
| − | 22 28 1 0 0 0 0 | + | 22 28 1 0 0 0 0 |
| − | 27 29 1 0 0 0 0 | + | 27 29 1 0 0 0 0 |
| − | 26 30 1 0 0 0 0 | + | 26 30 1 0 0 0 0 |
| − | 23 19 1 0 0 0 0 | + | 23 19 1 0 0 0 0 |
| − | 31 32 1 1 0 0 0 | + | 31 32 1 1 0 0 0 |
| − | 32 33 1 1 0 0 0 | + | 32 33 1 1 0 0 0 |
| − | 34 33 1 1 0 0 0 | + | 34 33 1 1 0 0 0 |
| − | 34 35 1 0 0 0 0 | + | 34 35 1 0 0 0 0 |
| − | 35 36 1 0 0 0 0 | + | 35 36 1 0 0 0 0 |
| − | 36 31 1 0 0 0 0 | + | 36 31 1 0 0 0 0 |
| − | 32 37 1 0 0 0 0 | + | 32 37 1 0 0 0 0 |
| − | 33 38 1 0 0 0 0 | + | 33 38 1 0 0 0 0 |
| − | 28 31 1 0 0 0 0 | + | 28 31 1 0 0 0 0 |
| − | 34 39 1 0 0 0 0 | + | 34 39 1 0 0 0 0 |
| − | 16 40 1 0 0 0 0 | + | 16 40 1 0 0 0 0 |
| − | 42 41 1 1 0 0 0 | + | 42 41 1 1 0 0 0 |
| − | 42 43 1 0 0 0 0 | + | 42 43 1 0 0 0 0 |
| − | 43 44 1 0 0 0 0 | + | 43 44 1 0 0 0 0 |
| − | 44 45 1 0 0 0 0 | + | 44 45 1 0 0 0 0 |
| − | 45 46 1 1 0 0 0 | + | 45 46 1 1 0 0 0 |
| − | 46 41 1 1 0 0 0 | + | 46 41 1 1 0 0 0 |
| − | 45 47 1 0 0 0 0 | + | 45 47 1 0 0 0 0 |
| − | 44 48 1 0 0 0 0 | + | 44 48 1 0 0 0 0 |
| − | 46 49 1 0 0 0 0 | + | 46 49 1 0 0 0 0 |
| − | 41 50 1 0 0 0 0 | + | 41 50 1 0 0 0 0 |
| − | 42 18 1 0 0 0 0 | + | 42 18 1 0 0 0 0 |
| − | 51 52 1 0 0 0 0 | + | 51 52 1 0 0 0 0 |
| − | 35 51 1 0 0 0 0 | + | 35 51 1 0 0 0 0 |
| − | 53 54 1 0 0 0 0 | + | 53 54 1 0 0 0 0 |
| − | 25 53 1 0 0 0 0 | + | 25 53 1 0 0 0 0 |
| − | M STY 1 1 SUP | + | M STY 1 1 SUP |
| − | M SLB 1 1 1 | + | M SLB 1 1 1 |
| − | M SAL 1 2 51 52 | + | M SAL 1 2 51 52 |
| − | M SBL 1 1 57 | + | M SBL 1 1 57 |
| − | M SMT 1 CH2OH | + | M SMT 1 CH2OH |
| − | M SBV 1 57 -0.7284 -0.2241 | + | M SBV 1 57 -0.7284 -0.2241 |
| − | M STY 1 2 SUP | + | M STY 1 2 SUP |
| − | M SLB 1 2 2 | + | M SLB 1 2 2 |
| − | M SAL 2 2 53 54 | + | M SAL 2 2 53 54 |
| − | M SBL 2 1 59 | + | M SBL 2 1 59 |
| − | M SMT 2 ^ CH2OH | + | M SMT 2 ^ CH2OH |
| − | M SBV 2 59 0.1419 1.0660 | + | M SBV 2 59 0.1419 1.0660 |
| − | S SKP 5 | + | S SKP 5 |
| − | ID FL5FACGL0034 | + | ID FL5FACGL0034 |
| − | FORMULA C33H40O21 | + | FORMULA C33H40O21 |
| − | EXACTMASS 772.206208342 | + | EXACTMASS 772.206208342 |
| − | AVERAGEMASS 772.6581 | + | AVERAGEMASS 772.6581 |
| − | SMILES O(C(C)1)C(Oc(c6)cc(c(c56)C(C(=C(O5)c(c4)cc(c(c4)O)O)OC(O3)C(C(C(O)C3CO)O)OC(C2O)OC(CO)C(C2O)O)=O)O)C(O)C(O)C1O | + | SMILES O(C(C)1)C(Oc(c6)cc(c(c56)C(C(=C(O5)c(c4)cc(c(c4)O)O)OC(O3)C(C(C(O)C3CO)O)OC(C2O)OC(CO)C(C2O)O)=O)O)C(O)C(O)C1O |
M END | M END | ||
| − | |||
Latest revision as of 09:00, 14 March 2009
Copyright: ARM project http://www.metabolome.jp/
54 59 0 0 0 0 0 0 0 0999 V2000
-2.0380 0.7467 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.0380 -0.0629 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.3368 -0.4677 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.6357 -0.0629 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.6357 0.7467 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.3368 1.1515 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.0654 -0.4677 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.7665 -0.0629 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.7665 0.7467 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.0654 1.1515 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.0654 -1.0989 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.4674 1.1513 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.1818 0.7388 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.8964 1.1513 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.8964 1.9764 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.1818 2.3890 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.4674 1.9764 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.7388 1.1513 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.4103 -0.4527 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.3368 -1.2770 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.6953 2.4377 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.8380 -1.6111 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.1034 -1.1871 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.3366 -2.0025 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.1034 -2.8230 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.8380 -3.2471 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.6051 -2.4315 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.7727 -0.8868 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.2086 -2.8719 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.6644 -3.8168 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.5227 -0.3619 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.0803 -1.0979 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.8833 -0.7856 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.6580 -0.7773 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.0950 -0.2142 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.3926 -0.5848 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.4601 -1.0796 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
4.6540 -1.2706 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
5.0475 -0.5116 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.1818 3.2139 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.3800 1.4462 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.5882 0.9891 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.8395 1.8682 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.5882 2.7525 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.3800 3.2098 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.1289 2.3307 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.9727 3.8890 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.5689 3.4247 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.7778 2.7883 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-5.0475 1.4513 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
4.8234 0.0099 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.5871 1.2479 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.9615 -3.8890 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.2911 -3.7502 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1 2 1 0 0 0 0
2 3 2 0 0 0 0
3 4 1 0 0 0 0
4 5 2 0 0 0 0
5 6 1 0 0 0 0
6 1 2 0 0 0 0
4 7 1 0 0 0 0
7 8 1 0 0 0 0
8 9 2 0 0 0 0
9 10 1 0 0 0 0
10 5 1 0 0 0 0
7 11 2 0 0 0 0
9 12 1 0 0 0 0
12 13 2 0 0 0 0
13 14 1 0 0 0 0
14 15 2 0 0 0 0
15 16 1 0 0 0 0
16 17 2 0 0 0 0
17 12 1 0 0 0 0
1 18 1 0 0 0 0
8 19 1 0 0 0 0
3 20 1 0 0 0 0
21 15 1 0 0 0 0
22 23 1 0 0 0 0
23 24 1 1 0 0 0
24 25 1 1 0 0 0
26 25 1 1 0 0 0
26 27 1 0 0 0 0
27 22 1 0 0 0 0
22 28 1 0 0 0 0
27 29 1 0 0 0 0
26 30 1 0 0 0 0
23 19 1 0 0 0 0
31 32 1 1 0 0 0
32 33 1 1 0 0 0
34 33 1 1 0 0 0
34 35 1 0 0 0 0
35 36 1 0 0 0 0
36 31 1 0 0 0 0
32 37 1 0 0 0 0
33 38 1 0 0 0 0
28 31 1 0 0 0 0
34 39 1 0 0 0 0
16 40 1 0 0 0 0
42 41 1 1 0 0 0
42 43 1 0 0 0 0
43 44 1 0 0 0 0
44 45 1 0 0 0 0
45 46 1 1 0 0 0
46 41 1 1 0 0 0
45 47 1 0 0 0 0
44 48 1 0 0 0 0
46 49 1 0 0 0 0
41 50 1 0 0 0 0
42 18 1 0 0 0 0
51 52 1 0 0 0 0
35 51 1 0 0 0 0
53 54 1 0 0 0 0
25 53 1 0 0 0 0
M STY 1 1 SUP
M SLB 1 1 1
M SAL 1 2 51 52
M SBL 1 1 57
M SMT 1 CH2OH
M SBV 1 57 -0.7284 -0.2241
M STY 1 2 SUP
M SLB 1 2 2
M SAL 2 2 53 54
M SBL 2 1 59
M SMT 2 ^ CH2OH
M SBV 2 59 0.1419 1.0660
S SKP 5
ID FL5FACGL0034
FORMULA C33H40O21
EXACTMASS 772.206208342
AVERAGEMASS 772.6581
SMILES O(C(C)1)C(Oc(c6)cc(c(c56)C(C(=C(O5)c(c4)cc(c(c4)O)O)OC(O3)C(C(C(O)C3CO)O)OC(C2O)OC(CO)C(C2O)O)=O)O)C(O)C(O)C1O
M END