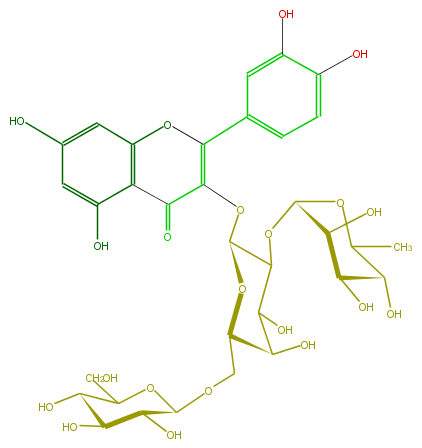
Mol:FL5FACGA0019
From Metabolomics.JP
(Difference between revisions)
| Line 1: | Line 1: | ||
| − | + | ||
| − | + | ||
| − | Copyright: ARM project http://www.metabolome.jp/ | + | Copyright: ARM project http://www.metabolome.jp/ |
| − | 54 59 0 0 0 0 0 0 0 0999 V2000 | + | 54 59 0 0 0 0 0 0 0 0999 V2000 |
| − | -2.9234 1.5915 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.9234 1.5915 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.9234 0.7820 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.9234 0.7820 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.2224 0.3774 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.2224 0.3774 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.5214 0.7820 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.5214 0.7820 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.5214 1.5915 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.5214 1.5915 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.2224 1.9963 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.2224 1.9963 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.8203 0.3774 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.8203 0.3774 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.1193 0.7820 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.1193 0.7820 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.1193 1.5915 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.1193 1.5915 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.8203 1.9963 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.8203 1.9963 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.8203 -0.2538 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.8203 -0.2538 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.7634 2.1520 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.7634 2.1520 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.4779 1.7396 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.4779 1.7396 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.1924 2.1520 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.1924 2.1520 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.1924 2.9771 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.1924 2.9771 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.4779 3.3895 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.4779 3.3895 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.7634 2.9771 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.7634 2.9771 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.2224 -0.4319 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.2224 -0.4319 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.9571 3.4186 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.9571 3.4186 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.6318 0.2866 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.6318 0.2866 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.4779 4.2143 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.4779 4.2143 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.7608 2.0750 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.7608 2.0750 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.6012 -1.0842 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.6012 -1.0842 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.5153 -1.0842 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.5153 -1.0842 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.8582 -0.4486 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.8582 -0.4486 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.6337 0.4429 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.6337 0.4429 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.7194 0.4429 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.7194 0.4429 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.3765 -0.1928 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.3765 -0.1928 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.2391 0.2500 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.2391 0.2500 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.0755 -1.6410 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.0755 -1.6410 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.6458 -1.7778 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.6458 -1.7778 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.7608 -0.4486 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.7608 -0.4486 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.2528 -0.8382 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.2528 -0.8382 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.4067 -0.3498 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.4067 -0.3498 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.6751 -1.2891 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.6751 -1.2891 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.4067 -2.2342 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.4067 -2.2342 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.2727 -2.6107 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.2727 -2.6107 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.9844 -1.7833 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.9844 -1.7833 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.1953 -0.1785 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.1953 -0.1785 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.4566 -2.0987 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.4566 -2.0987 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.9276 -2.3964 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.9276 -2.3964 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.5112 -2.9869 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.5112 -2.9869 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.0005 -3.3243 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.0005 -3.3243 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.5932 -3.3953 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.5932 -3.3953 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.0870 -4.0634 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.0870 -4.0634 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.3581 -3.7800 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.3581 -3.7800 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.6547 -3.7723 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.6547 -3.7723 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.1658 -3.2611 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.1658 -3.2611 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.8036 -3.5977 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.8036 -3.5977 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.1895 -3.6362 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.1895 -3.6362 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.7097 -4.0413 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.7097 -4.0413 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.7534 -4.2143 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.7534 -4.2143 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.3970 -3.0557 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.3970 -3.0557 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.4012 -2.7867 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.4012 -2.7867 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1 2 1 0 0 0 0 | + | 1 2 1 0 0 0 0 |
| − | 2 3 2 0 0 0 0 | + | 2 3 2 0 0 0 0 |
| − | 4 5 2 0 0 0 0 | + | 4 5 2 0 0 0 0 |
| − | 5 6 1 0 0 0 0 | + | 5 6 1 0 0 0 0 |
| − | 6 1 2 0 0 0 0 | + | 6 1 2 0 0 0 0 |
| − | 4 7 1 0 0 0 0 | + | 4 7 1 0 0 0 0 |
| − | 7 8 1 0 0 0 0 | + | 7 8 1 0 0 0 0 |
| − | 8 9 2 0 0 0 0 | + | 8 9 2 0 0 0 0 |
| − | 9 10 1 0 0 0 0 | + | 9 10 1 0 0 0 0 |
| − | 10 5 1 0 0 0 0 | + | 10 5 1 0 0 0 0 |
| − | 7 11 2 0 0 0 0 | + | 7 11 2 0 0 0 0 |
| − | 9 12 1 0 0 0 0 | + | 9 12 1 0 0 0 0 |
| − | 12 13 2 0 0 0 0 | + | 12 13 2 0 0 0 0 |
| − | 13 14 1 0 0 0 0 | + | 13 14 1 0 0 0 0 |
| − | 14 15 2 0 0 0 0 | + | 14 15 2 0 0 0 0 |
| − | 15 16 1 0 0 0 0 | + | 15 16 1 0 0 0 0 |
| − | 16 17 2 0 0 0 0 | + | 16 17 2 0 0 0 0 |
| − | 17 12 1 0 0 0 0 | + | 17 12 1 0 0 0 0 |
| − | 3 18 1 0 0 0 0 | + | 3 18 1 0 0 0 0 |
| − | 19 15 1 0 0 0 0 | + | 19 15 1 0 0 0 0 |
| − | 20 8 1 0 0 0 0 | + | 20 8 1 0 0 0 0 |
| − | 4 3 1 0 0 0 0 | + | 4 3 1 0 0 0 0 |
| − | 16 21 1 0 0 0 0 | + | 16 21 1 0 0 0 0 |
| − | 1 22 1 0 0 0 0 | + | 1 22 1 0 0 0 0 |
| − | 24 23 1 1 0 0 0 | + | 24 23 1 1 0 0 0 |
| − | 24 25 1 0 0 0 0 | + | 24 25 1 0 0 0 0 |
| − | 25 26 1 0 0 0 0 | + | 25 26 1 0 0 0 0 |
| − | 26 27 1 0 0 0 0 | + | 26 27 1 0 0 0 0 |
| − | 27 28 1 1 0 0 0 | + | 27 28 1 1 0 0 0 |
| − | 28 23 1 1 0 0 0 | + | 28 23 1 1 0 0 0 |
| − | 28 29 1 0 0 0 0 | + | 28 29 1 0 0 0 0 |
| − | 23 30 1 0 0 0 0 | + | 23 30 1 0 0 0 0 |
| − | 24 31 1 0 0 0 0 | + | 24 31 1 0 0 0 0 |
| − | 25 32 1 0 0 0 0 | + | 25 32 1 0 0 0 0 |
| − | 33 34 1 0 0 0 0 | + | 33 34 1 0 0 0 0 |
| − | 34 35 1 1 0 0 0 | + | 34 35 1 1 0 0 0 |
| − | 35 36 1 1 0 0 0 | + | 35 36 1 1 0 0 0 |
| − | 37 36 1 1 0 0 0 | + | 37 36 1 1 0 0 0 |
| − | 37 38 1 0 0 0 0 | + | 37 38 1 0 0 0 0 |
| − | 38 33 1 0 0 0 0 | + | 38 33 1 0 0 0 0 |
| − | 33 39 1 0 0 0 0 | + | 33 39 1 0 0 0 0 |
| − | 38 40 1 0 0 0 0 | + | 38 40 1 0 0 0 0 |
| − | 37 41 1 0 0 0 0 | + | 37 41 1 0 0 0 0 |
| − | 36 42 1 0 0 0 0 | + | 36 42 1 0 0 0 0 |
| − | 34 20 1 0 0 0 0 | + | 34 20 1 0 0 0 0 |
| − | 27 39 1 0 0 0 0 | + | 27 39 1 0 0 0 0 |
| − | 43 42 1 0 0 0 0 | + | 43 42 1 0 0 0 0 |
| − | 44 45 1 1 0 0 0 | + | 44 45 1 1 0 0 0 |
| − | 45 46 1 1 0 0 0 | + | 45 46 1 1 0 0 0 |
| − | 47 46 1 1 0 0 0 | + | 47 46 1 1 0 0 0 |
| − | 47 48 1 0 0 0 0 | + | 47 48 1 0 0 0 0 |
| − | 48 49 1 0 0 0 0 | + | 48 49 1 0 0 0 0 |
| − | 49 44 1 0 0 0 0 | + | 49 44 1 0 0 0 0 |
| − | 44 50 1 0 0 0 0 | + | 44 50 1 0 0 0 0 |
| − | 45 51 1 0 0 0 0 | + | 45 51 1 0 0 0 0 |
| − | 46 52 1 0 0 0 0 | + | 46 52 1 0 0 0 0 |
| − | 47 43 1 0 0 0 0 | + | 47 43 1 0 0 0 0 |
| − | 53 54 1 0 0 0 0 | + | 53 54 1 0 0 0 0 |
| − | 49 53 1 0 0 0 0 | + | 49 53 1 0 0 0 0 |
| − | M STY 1 1 SUP | + | M STY 1 1 SUP |
| − | M SLB 1 1 1 | + | M SLB 1 1 1 |
| − | M SAL 1 2 53 54 | + | M SAL 1 2 53 54 |
| − | M SBL 1 1 59 | + | M SBL 1 1 59 |
| − | M SMT 1 ^CH2OH | + | M SMT 1 ^CH2OH |
| − | M SBV 1 59 0.5934 -0.5419 | + | M SBV 1 59 0.5934 -0.5419 |
| − | S SKP 5 | + | S SKP 5 |
| − | ID FL5FACGA0019 | + | ID FL5FACGA0019 |
| − | FORMULA C33H40O21 | + | FORMULA C33H40O21 |
| − | EXACTMASS 772.206208342 | + | EXACTMASS 772.206208342 |
| − | AVERAGEMASS 772.6581 | + | AVERAGEMASS 772.6581 |
| − | SMILES O(C(CO)6)C(C(O)C(C(O)6)O)OCC(C(O)4)OC(C(OC(O5)C(C(O)C(C5C)O)O)C4O)OC(=C2c(c3)cc(c(c3)O)O)C(=O)c(c(O2)1)c(cc(O)c1)O | + | SMILES O(C(CO)6)C(C(O)C(C(O)6)O)OCC(C(O)4)OC(C(OC(O5)C(C(O)C(C5C)O)O)C4O)OC(=C2c(c3)cc(c(c3)O)O)C(=O)c(c(O2)1)c(cc(O)c1)O |
M END | M END | ||
| − | |||
Latest revision as of 09:00, 14 March 2009
Copyright: ARM project http://www.metabolome.jp/
54 59 0 0 0 0 0 0 0 0999 V2000
-2.9234 1.5915 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.9234 0.7820 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.2224 0.3774 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.5214 0.7820 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.5214 1.5915 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.2224 1.9963 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.8203 0.3774 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.1193 0.7820 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.1193 1.5915 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.8203 1.9963 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.8203 -0.2538 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.7634 2.1520 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.4779 1.7396 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.1924 2.1520 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.1924 2.9771 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.4779 3.3895 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.7634 2.9771 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.2224 -0.4319 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.9571 3.4186 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.6318 0.2866 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.4779 4.2143 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.7608 2.0750 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.6012 -1.0842 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.5153 -1.0842 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.8582 -0.4486 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.6337 0.4429 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.7194 0.4429 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.3765 -0.1928 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.2391 0.2500 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.0755 -1.6410 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.6458 -1.7778 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.7608 -0.4486 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.2528 -0.8382 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.4067 -0.3498 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.6751 -1.2891 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.4067 -2.2342 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.2727 -2.6107 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.9844 -1.7833 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.1953 -0.1785 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.4566 -2.0987 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.9276 -2.3964 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.5112 -2.9869 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.0005 -3.3243 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.5932 -3.3953 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.0870 -4.0634 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.3581 -3.7800 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.6547 -3.7723 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.1658 -3.2611 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.8036 -3.5977 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.1895 -3.6362 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.7097 -4.0413 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.7534 -4.2143 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.3970 -3.0557 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.4012 -2.7867 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1 2 1 0 0 0 0
2 3 2 0 0 0 0
4 5 2 0 0 0 0
5 6 1 0 0 0 0
6 1 2 0 0 0 0
4 7 1 0 0 0 0
7 8 1 0 0 0 0
8 9 2 0 0 0 0
9 10 1 0 0 0 0
10 5 1 0 0 0 0
7 11 2 0 0 0 0
9 12 1 0 0 0 0
12 13 2 0 0 0 0
13 14 1 0 0 0 0
14 15 2 0 0 0 0
15 16 1 0 0 0 0
16 17 2 0 0 0 0
17 12 1 0 0 0 0
3 18 1 0 0 0 0
19 15 1 0 0 0 0
20 8 1 0 0 0 0
4 3 1 0 0 0 0
16 21 1 0 0 0 0
1 22 1 0 0 0 0
24 23 1 1 0 0 0
24 25 1 0 0 0 0
25 26 1 0 0 0 0
26 27 1 0 0 0 0
27 28 1 1 0 0 0
28 23 1 1 0 0 0
28 29 1 0 0 0 0
23 30 1 0 0 0 0
24 31 1 0 0 0 0
25 32 1 0 0 0 0
33 34 1 0 0 0 0
34 35 1 1 0 0 0
35 36 1 1 0 0 0
37 36 1 1 0 0 0
37 38 1 0 0 0 0
38 33 1 0 0 0 0
33 39 1 0 0 0 0
38 40 1 0 0 0 0
37 41 1 0 0 0 0
36 42 1 0 0 0 0
34 20 1 0 0 0 0
27 39 1 0 0 0 0
43 42 1 0 0 0 0
44 45 1 1 0 0 0
45 46 1 1 0 0 0
47 46 1 1 0 0 0
47 48 1 0 0 0 0
48 49 1 0 0 0 0
49 44 1 0 0 0 0
44 50 1 0 0 0 0
45 51 1 0 0 0 0
46 52 1 0 0 0 0
47 43 1 0 0 0 0
53 54 1 0 0 0 0
49 53 1 0 0 0 0
M STY 1 1 SUP
M SLB 1 1 1
M SAL 1 2 53 54
M SBL 1 1 59
M SMT 1 ^CH2OH
M SBV 1 59 0.5934 -0.5419
S SKP 5
ID FL5FACGA0019
FORMULA C33H40O21
EXACTMASS 772.206208342
AVERAGEMASS 772.6581
SMILES O(C(CO)6)C(C(O)C(C(O)6)O)OCC(C(O)4)OC(C(OC(O5)C(C(O)C(C5C)O)O)C4O)OC(=C2c(c3)cc(c(c3)O)O)C(=O)c(c(O2)1)c(cc(O)c1)O
M END