Mol:FL5FAAGA0029
From Metabolomics.JP
(Difference between revisions)
| Line 1: | Line 1: | ||
| − | + | ||
| − | + | ||
| − | Copyright: ARM project http://www.metabolome.jp/ | + | Copyright: ARM project http://www.metabolome.jp/ |
| − | 43 47 0 0 0 0 0 0 0 0999 V2000 | + | 43 47 0 0 0 0 0 0 0 0999 V2000 |
| − | -2.6264 0.6689 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.6264 0.6689 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.0554 0.9986 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.0554 0.9986 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.0554 1.6580 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.0554 1.6580 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.6264 1.9877 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.6264 1.9877 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.1975 1.6580 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.1975 1.6580 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.1975 0.9986 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.1975 0.9986 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.4843 0.6689 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.4843 0.6689 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.9133 0.9986 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.9133 0.9986 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.9133 1.6580 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.9133 1.6580 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.4843 1.9877 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.4843 1.9877 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.3676 1.9731 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.3676 1.9731 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.2028 1.6437 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.2028 1.6437 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.7733 1.9731 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.7733 1.9731 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.7733 2.6318 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.7733 2.6318 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.2028 2.9611 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.2028 2.9611 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.3676 2.6318 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.3676 2.6318 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.2203 2.8899 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.2203 2.8899 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.5797 1.8787 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.5797 1.8787 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.6264 0.1904 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.6264 0.1904 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.4843 0.2894 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.4843 0.2894 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.3547 0.6761 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.3547 0.6761 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.2181 -0.0241 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.2181 -0.0241 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.7573 0.2873 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.7573 0.2873 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.1533 0.4384 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.1533 0.4384 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.7106 0.8763 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.7106 0.8763 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.1713 0.5650 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.1713 0.5650 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.7754 0.4138 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.7754 0.4138 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.2605 0.8763 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.2605 0.8763 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.4659 0.7511 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.4659 0.7511 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.6982 -0.3013 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.6982 -0.3013 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.9429 0.6087 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.9429 0.6087 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.6982 -0.8493 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.6982 -0.8493 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.6982 -1.3615 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.6982 -1.3615 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.3323 -1.5728 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.3323 -1.5728 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.0422 -1.5601 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.0422 -1.5601 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.0422 -2.2023 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.0422 -2.2023 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.5983 -2.5233 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.5983 -2.5233 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.1544 -2.2023 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.1544 -2.2023 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.1544 -1.5601 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.1544 -1.5601 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.5983 -1.2391 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.5983 -1.2391 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.5797 -1.3146 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.5797 -1.3146 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.5742 -2.4446 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.5742 -2.4446 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.5983 -2.9611 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.5983 -2.9611 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1 2 1 0 0 0 0 | + | 1 2 1 0 0 0 0 |
| − | 2 3 2 0 0 0 0 | + | 2 3 2 0 0 0 0 |
| − | 3 4 1 0 0 0 0 | + | 3 4 1 0 0 0 0 |
| − | 4 5 2 0 0 0 0 | + | 4 5 2 0 0 0 0 |
| − | 5 6 1 0 0 0 0 | + | 5 6 1 0 0 0 0 |
| − | 6 1 2 0 0 0 0 | + | 6 1 2 0 0 0 0 |
| − | 2 7 1 0 0 0 0 | + | 2 7 1 0 0 0 0 |
| − | 7 8 1 0 0 0 0 | + | 7 8 1 0 0 0 0 |
| − | 8 9 2 0 0 0 0 | + | 8 9 2 0 0 0 0 |
| − | 9 10 1 0 0 0 0 | + | 9 10 1 0 0 0 0 |
| − | 10 3 1 0 0 0 0 | + | 10 3 1 0 0 0 0 |
| − | 9 11 1 0 0 0 0 | + | 9 11 1 0 0 0 0 |
| − | 11 12 2 0 0 0 0 | + | 11 12 2 0 0 0 0 |
| − | 12 13 1 0 0 0 0 | + | 12 13 1 0 0 0 0 |
| − | 13 14 2 0 0 0 0 | + | 13 14 2 0 0 0 0 |
| − | 14 15 1 0 0 0 0 | + | 14 15 1 0 0 0 0 |
| − | 15 16 2 0 0 0 0 | + | 15 16 2 0 0 0 0 |
| − | 16 11 1 0 0 0 0 | + | 16 11 1 0 0 0 0 |
| − | 14 17 1 0 0 0 0 | + | 14 17 1 0 0 0 0 |
| − | 5 18 1 0 0 0 0 | + | 5 18 1 0 0 0 0 |
| − | 1 19 1 0 0 0 0 | + | 1 19 1 0 0 0 0 |
| − | 7 20 2 0 0 0 0 | + | 7 20 2 0 0 0 0 |
| − | 8 21 1 0 0 0 0 | + | 8 21 1 0 0 0 0 |
| − | 23 22 1 1 0 0 0 | + | 23 22 1 1 0 0 0 |
| − | 23 24 1 0 0 0 0 | + | 23 24 1 0 0 0 0 |
| − | 24 25 1 0 0 0 0 | + | 24 25 1 0 0 0 0 |
| − | 25 26 1 0 0 0 0 | + | 25 26 1 0 0 0 0 |
| − | 26 27 1 1 0 0 0 | + | 26 27 1 1 0 0 0 |
| − | 22 27 1 1 0 0 0 | + | 22 27 1 1 0 0 0 |
| − | 21 26 1 0 0 0 0 | + | 21 26 1 0 0 0 0 |
| − | 25 28 1 0 0 0 0 | + | 25 28 1 0 0 0 0 |
| − | 24 29 1 0 0 0 0 | + | 24 29 1 0 0 0 0 |
| − | 22 30 1 0 0 0 0 | + | 22 30 1 0 0 0 0 |
| − | 23 31 1 0 0 0 0 | + | 23 31 1 0 0 0 0 |
| − | 30 32 1 0 0 0 0 | + | 30 32 1 0 0 0 0 |
| − | 32 33 1 0 0 0 0 | + | 32 33 1 0 0 0 0 |
| − | 33 34 2 0 0 0 0 | + | 33 34 2 0 0 0 0 |
| − | 33 35 1 0 0 0 0 | + | 33 35 1 0 0 0 0 |
| − | 35 36 1 0 0 0 0 | + | 35 36 1 0 0 0 0 |
| − | 36 37 2 0 0 0 0 | + | 36 37 2 0 0 0 0 |
| − | 37 38 1 0 0 0 0 | + | 37 38 1 0 0 0 0 |
| − | 38 39 2 0 0 0 0 | + | 38 39 2 0 0 0 0 |
| − | 39 40 1 0 0 0 0 | + | 39 40 1 0 0 0 0 |
| − | 40 35 2 0 0 0 0 | + | 40 35 2 0 0 0 0 |
| − | 39 41 1 0 0 0 0 | + | 39 41 1 0 0 0 0 |
| − | 38 42 1 0 0 0 0 | + | 38 42 1 0 0 0 0 |
| − | 37 43 1 0 0 0 0 | + | 37 43 1 0 0 0 0 |
| − | S SKP 8 | + | S SKP 8 |
| − | ID FL5FAAGA0029 | + | ID FL5FAAGA0029 |
| − | KNApSAcK_ID C00005841 | + | KNApSAcK_ID C00005841 |
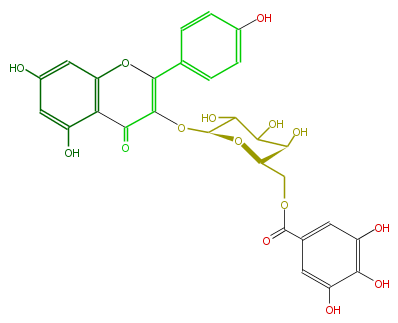
| − | NAME Kaempferol 3-(6''-galloylgalactoside);5,7-Dihydroxy-2-(4-hydroxyphenyl)-3-[[6-O-(3,4,5-trihydroxybenzoyl)-beta-D-galactopyranosyl]oxy]-4H-1-benzopyran-4-one | + | NAME Kaempferol 3-(6''-galloylgalactoside);5,7-Dihydroxy-2-(4-hydroxyphenyl)-3-[[6-O-(3,4,5-trihydroxybenzoyl)-beta-D-galactopyranosyl]oxy]-4H-1-benzopyran-4-one |
| − | CAS_RN 56317-06-7 | + | CAS_RN 56317-06-7 |
| − | FORMULA C28H24O15 | + | FORMULA C28H24O15 |
| − | EXACTMASS 600.111520098 | + | EXACTMASS 600.111520098 |
| − | AVERAGEMASS 600.48116 | + | AVERAGEMASS 600.48116 |
| − | SMILES Oc(c1)c(c(O)cc1C(OCC(C(O)5)OC(C(O)C5O)OC(=C3c(c4)ccc(O)c4)C(c(c(O3)2)c(cc(O)c2)O)=O)=O)O | + | SMILES Oc(c1)c(c(O)cc1C(OCC(C(O)5)OC(C(O)C5O)OC(=C3c(c4)ccc(O)c4)C(c(c(O3)2)c(cc(O)c2)O)=O)=O)O |
M END | M END | ||
| − | |||
Latest revision as of 09:00, 14 March 2009
Copyright: ARM project http://www.metabolome.jp/
43 47 0 0 0 0 0 0 0 0999 V2000
-2.6264 0.6689 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.0554 0.9986 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.0554 1.6580 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.6264 1.9877 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.1975 1.6580 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.1975 0.9986 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.4843 0.6689 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.9133 0.9986 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.9133 1.6580 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.4843 1.9877 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.3676 1.9731 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.2028 1.6437 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.7733 1.9731 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.7733 2.6318 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.2028 2.9611 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.3676 2.6318 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.2203 2.8899 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.5797 1.8787 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.6264 0.1904 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.4843 0.2894 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.3547 0.6761 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.2181 -0.0241 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.7573 0.2873 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.1533 0.4384 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.7106 0.8763 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.1713 0.5650 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.7754 0.4138 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.2605 0.8763 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.4659 0.7511 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.6982 -0.3013 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.9429 0.6087 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.6982 -0.8493 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.6982 -1.3615 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.3323 -1.5728 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.0422 -1.5601 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.0422 -2.2023 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.5983 -2.5233 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.1544 -2.2023 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.1544 -1.5601 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.5983 -1.2391 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.5797 -1.3146 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.5742 -2.4446 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.5983 -2.9611 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1 2 1 0 0 0 0
2 3 2 0 0 0 0
3 4 1 0 0 0 0
4 5 2 0 0 0 0
5 6 1 0 0 0 0
6 1 2 0 0 0 0
2 7 1 0 0 0 0
7 8 1 0 0 0 0
8 9 2 0 0 0 0
9 10 1 0 0 0 0
10 3 1 0 0 0 0
9 11 1 0 0 0 0
11 12 2 0 0 0 0
12 13 1 0 0 0 0
13 14 2 0 0 0 0
14 15 1 0 0 0 0
15 16 2 0 0 0 0
16 11 1 0 0 0 0
14 17 1 0 0 0 0
5 18 1 0 0 0 0
1 19 1 0 0 0 0
7 20 2 0 0 0 0
8 21 1 0 0 0 0
23 22 1 1 0 0 0
23 24 1 0 0 0 0
24 25 1 0 0 0 0
25 26 1 0 0 0 0
26 27 1 1 0 0 0
22 27 1 1 0 0 0
21 26 1 0 0 0 0
25 28 1 0 0 0 0
24 29 1 0 0 0 0
22 30 1 0 0 0 0
23 31 1 0 0 0 0
30 32 1 0 0 0 0
32 33 1 0 0 0 0
33 34 2 0 0 0 0
33 35 1 0 0 0 0
35 36 1 0 0 0 0
36 37 2 0 0 0 0
37 38 1 0 0 0 0
38 39 2 0 0 0 0
39 40 1 0 0 0 0
40 35 2 0 0 0 0
39 41 1 0 0 0 0
38 42 1 0 0 0 0
37 43 1 0 0 0 0
S SKP 8
ID FL5FAAGA0029
KNApSAcK_ID C00005841
NAME Kaempferol 3-(6''-galloylgalactoside);5,7-Dihydroxy-2-(4-hydroxyphenyl)-3-[[6-O-(3,4,5-trihydroxybenzoyl)-beta-D-galactopyranosyl]oxy]-4H-1-benzopyran-4-one
CAS_RN 56317-06-7
FORMULA C28H24O15
EXACTMASS 600.111520098
AVERAGEMASS 600.48116
SMILES Oc(c1)c(c(O)cc1C(OCC(C(O)5)OC(C(O)C5O)OC(=C3c(c4)ccc(O)c4)C(c(c(O3)2)c(cc(O)c2)O)=O)=O)O
M END