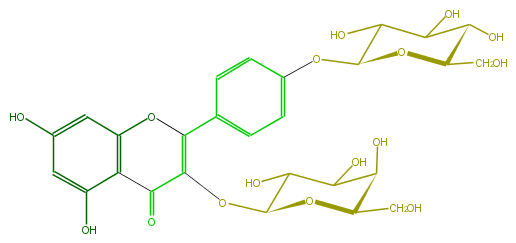
Mol:FL5FAAGA0012
From Metabolomics.JP
(Difference between revisions)
| Line 1: | Line 1: | ||
| − | + | ||
| − | + | ||
| − | Copyright: ARM project http://www.metabolome.jp/ | + | Copyright: ARM project http://www.metabolome.jp/ |
| − | 43 47 0 0 0 0 0 0 0 0999 V2000 | + | 43 47 0 0 0 0 0 0 0 0999 V2000 |
| − | -4.6681 -0.2737 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.6681 -0.2737 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.6681 -1.0832 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.6681 -1.0832 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.9671 -1.4879 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.9671 -1.4879 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.2661 -1.0832 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.2661 -1.0832 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.2661 -0.2737 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.2661 -0.2737 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.9671 0.1310 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.9671 0.1310 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.5649 -1.4879 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.5649 -1.4879 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.8639 -1.0832 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.8639 -1.0832 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.8639 -0.2737 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.8639 -0.2737 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.5649 0.1310 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.5649 0.1310 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.5649 -2.1191 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.5649 -2.1191 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.1633 0.1309 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.1633 0.1309 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.4488 -0.2817 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.4488 -0.2817 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.2657 0.1309 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.2657 0.1309 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.2657 0.9559 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.2657 0.9559 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.4488 1.3684 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.4488 1.3684 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.1633 0.9559 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.1633 0.9559 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.9671 -2.2971 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.9671 -2.2971 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -5.3689 0.1309 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -5.3689 0.1309 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.9800 1.3683 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.9800 1.3683 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.0212 -1.7246 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.0212 -1.7246 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.3906 -1.0971 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.3906 -1.0971 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.0978 -1.9433 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.0978 -1.9433 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.8417 -1.6748 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.8417 -1.6748 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.7866 -1.9433 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.7866 -1.9433 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.2753 -1.0971 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.2753 -1.0971 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.3358 -1.3655 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.3358 -1.3655 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.3025 -1.2930 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.3025 -1.2930 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.8285 -0.8428 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.8285 -0.8428 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.2767 -0.4084 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.2767 -0.4084 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.3717 2.0923 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.3717 2.0923 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.8831 1.2462 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.8831 1.2462 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.8226 1.5146 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.8226 1.5146 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.7677 1.2462 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.7677 1.2462 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.2562 2.0923 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.2562 2.0923 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.3168 1.8239 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.3168 1.8239 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.5276 1.8891 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.5276 1.8891 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.8287 2.3480 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.8287 2.3480 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.7769 1.8489 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.7769 1.8489 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.6255 -1.8282 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.6255 -1.8282 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.5258 -2.3480 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.5258 -2.3480 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.4685 1.2940 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.4685 1.2940 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 5.3689 0.7742 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 5.3689 0.7742 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1 2 1 0 0 0 0 | + | 1 2 1 0 0 0 0 |
| − | 2 3 2 0 0 0 0 | + | 2 3 2 0 0 0 0 |
| − | 3 4 1 0 0 0 0 | + | 3 4 1 0 0 0 0 |
| − | 4 5 2 0 0 0 0 | + | 4 5 2 0 0 0 0 |
| − | 5 6 1 0 0 0 0 | + | 5 6 1 0 0 0 0 |
| − | 6 1 2 0 0 0 0 | + | 6 1 2 0 0 0 0 |
| − | 4 7 1 0 0 0 0 | + | 4 7 1 0 0 0 0 |
| − | 7 8 1 0 0 0 0 | + | 7 8 1 0 0 0 0 |
| − | 8 9 2 0 0 0 0 | + | 8 9 2 0 0 0 0 |
| − | 9 10 1 0 0 0 0 | + | 9 10 1 0 0 0 0 |
| − | 10 5 1 0 0 0 0 | + | 10 5 1 0 0 0 0 |
| − | 7 11 2 0 0 0 0 | + | 7 11 2 0 0 0 0 |
| − | 9 12 1 0 0 0 0 | + | 9 12 1 0 0 0 0 |
| − | 12 13 2 0 0 0 0 | + | 12 13 2 0 0 0 0 |
| − | 13 14 1 0 0 0 0 | + | 13 14 1 0 0 0 0 |
| − | 14 15 2 0 0 0 0 | + | 14 15 2 0 0 0 0 |
| − | 15 16 1 0 0 0 0 | + | 15 16 1 0 0 0 0 |
| − | 16 17 2 0 0 0 0 | + | 16 17 2 0 0 0 0 |
| − | 17 12 1 0 0 0 0 | + | 17 12 1 0 0 0 0 |
| − | 3 18 1 0 0 0 0 | + | 3 18 1 0 0 0 0 |
| − | 1 19 1 0 0 0 0 | + | 1 19 1 0 0 0 0 |
| − | 15 20 1 0 0 0 0 | + | 15 20 1 0 0 0 0 |
| − | 8 21 1 0 0 0 0 | + | 8 21 1 0 0 0 0 |
| − | 22 23 1 0 0 0 0 | + | 22 23 1 0 0 0 0 |
| − | 23 24 1 1 0 0 0 | + | 23 24 1 1 0 0 0 |
| − | 24 25 1 1 0 0 0 | + | 24 25 1 1 0 0 0 |
| − | 26 25 1 1 0 0 0 | + | 26 25 1 1 0 0 0 |
| − | 26 27 1 0 0 0 0 | + | 26 27 1 0 0 0 0 |
| − | 27 22 1 0 0 0 0 | + | 27 22 1 0 0 0 0 |
| − | 22 28 1 0 0 0 0 | + | 22 28 1 0 0 0 0 |
| − | 27 29 1 0 0 0 0 | + | 27 29 1 0 0 0 0 |
| − | 26 30 1 0 0 0 0 | + | 26 30 1 0 0 0 0 |
| − | 23 21 1 0 0 0 0 | + | 23 21 1 0 0 0 0 |
| − | 31 32 1 0 0 0 0 | + | 31 32 1 0 0 0 0 |
| − | 32 33 1 1 0 0 0 | + | 32 33 1 1 0 0 0 |
| − | 33 34 1 1 0 0 0 | + | 33 34 1 1 0 0 0 |
| − | 35 34 1 1 0 0 0 | + | 35 34 1 1 0 0 0 |
| − | 35 36 1 0 0 0 0 | + | 35 36 1 0 0 0 0 |
| − | 36 31 1 0 0 0 0 | + | 36 31 1 0 0 0 0 |
| − | 31 37 1 0 0 0 0 | + | 31 37 1 0 0 0 0 |
| − | 36 38 1 0 0 0 0 | + | 36 38 1 0 0 0 0 |
| − | 35 39 1 0 0 0 0 | + | 35 39 1 0 0 0 0 |
| − | 32 20 1 0 0 0 0 | + | 32 20 1 0 0 0 0 |
| − | 40 41 1 0 0 0 0 | + | 40 41 1 0 0 0 0 |
| − | 25 40 1 0 0 0 0 | + | 25 40 1 0 0 0 0 |
| − | 42 43 1 0 0 0 0 | + | 42 43 1 0 0 0 0 |
| − | 34 42 1 0 0 0 0 | + | 34 42 1 0 0 0 0 |
| − | M STY 1 1 SUP | + | M STY 1 1 SUP |
| − | M SLB 1 1 1 | + | M SLB 1 1 1 |
| − | M SAL 1 2 40 41 | + | M SAL 1 2 40 41 |
| − | M SBL 1 1 45 | + | M SBL 1 1 45 |
| − | M SMT 1 CH2OH | + | M SMT 1 CH2OH |
| − | M SBV 1 45 -0.8388 -0.1151 | + | M SBV 1 45 -0.8388 -0.1151 |
| − | M STY 1 2 SUP | + | M STY 1 2 SUP |
| − | M SLB 1 2 2 | + | M SLB 1 2 2 |
| − | M SAL 2 2 42 43 | + | M SAL 2 2 42 43 |
| − | M SBL 2 1 47 | + | M SBL 2 1 47 |
| − | M SMT 2 CH2OH | + | M SMT 2 CH2OH |
| − | M SBV 2 47 -0.7008 -0.0478 | + | M SBV 2 47 -0.7008 -0.0478 |
| − | S SKP 5 | + | S SKP 5 |
| − | ID FL5FAAGA0012 | + | ID FL5FAAGA0012 |
| − | FORMULA C27H30O16 | + | FORMULA C27H30O16 |
| − | EXACTMASS 610.153384912 | + | EXACTMASS 610.153384912 |
| − | AVERAGEMASS 610.5175 | + | AVERAGEMASS 610.5175 |
| − | SMILES c(c1)(O3)c(C(C(=C(c(c4)ccc(OC(O5)C(O)C(O)C(O)C5CO)c4)3)OC(C(O)2)OC(C(C(O)2)O)CO)=O)c(O)cc(O)1 | + | SMILES c(c1)(O3)c(C(C(=C(c(c4)ccc(OC(O5)C(O)C(O)C(O)C5CO)c4)3)OC(C(O)2)OC(C(C(O)2)O)CO)=O)c(O)cc(O)1 |
M END | M END | ||
| − | |||
Latest revision as of 09:00, 14 March 2009
Copyright: ARM project http://www.metabolome.jp/
43 47 0 0 0 0 0 0 0 0999 V2000
-4.6681 -0.2737 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.6681 -1.0832 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.9671 -1.4879 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.2661 -1.0832 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.2661 -0.2737 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.9671 0.1310 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.5649 -1.4879 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.8639 -1.0832 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.8639 -0.2737 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.5649 0.1310 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.5649 -2.1191 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.1633 0.1309 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.4488 -0.2817 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.2657 0.1309 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.2657 0.9559 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.4488 1.3684 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.1633 0.9559 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.9671 -2.2971 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-5.3689 0.1309 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.9800 1.3683 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.0212 -1.7246 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.3906 -1.0971 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.0978 -1.9433 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.8417 -1.6748 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.7866 -1.9433 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.2753 -1.0971 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.3358 -1.3655 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.3025 -1.2930 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.8285 -0.8428 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.2767 -0.4084 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.3717 2.0923 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.8831 1.2462 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.8226 1.5146 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.7677 1.2462 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.2562 2.0923 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.3168 1.8239 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.5276 1.8891 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.8287 2.3480 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
4.7769 1.8489 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.6255 -1.8282 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.5258 -2.3480 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
4.4685 1.2940 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
5.3689 0.7742 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1 2 1 0 0 0 0
2 3 2 0 0 0 0
3 4 1 0 0 0 0
4 5 2 0 0 0 0
5 6 1 0 0 0 0
6 1 2 0 0 0 0
4 7 1 0 0 0 0
7 8 1 0 0 0 0
8 9 2 0 0 0 0
9 10 1 0 0 0 0
10 5 1 0 0 0 0
7 11 2 0 0 0 0
9 12 1 0 0 0 0
12 13 2 0 0 0 0
13 14 1 0 0 0 0
14 15 2 0 0 0 0
15 16 1 0 0 0 0
16 17 2 0 0 0 0
17 12 1 0 0 0 0
3 18 1 0 0 0 0
1 19 1 0 0 0 0
15 20 1 0 0 0 0
8 21 1 0 0 0 0
22 23 1 0 0 0 0
23 24 1 1 0 0 0
24 25 1 1 0 0 0
26 25 1 1 0 0 0
26 27 1 0 0 0 0
27 22 1 0 0 0 0
22 28 1 0 0 0 0
27 29 1 0 0 0 0
26 30 1 0 0 0 0
23 21 1 0 0 0 0
31 32 1 0 0 0 0
32 33 1 1 0 0 0
33 34 1 1 0 0 0
35 34 1 1 0 0 0
35 36 1 0 0 0 0
36 31 1 0 0 0 0
31 37 1 0 0 0 0
36 38 1 0 0 0 0
35 39 1 0 0 0 0
32 20 1 0 0 0 0
40 41 1 0 0 0 0
25 40 1 0 0 0 0
42 43 1 0 0 0 0
34 42 1 0 0 0 0
M STY 1 1 SUP
M SLB 1 1 1
M SAL 1 2 40 41
M SBL 1 1 45
M SMT 1 CH2OH
M SBV 1 45 -0.8388 -0.1151
M STY 1 2 SUP
M SLB 1 2 2
M SAL 2 2 42 43
M SBL 2 1 47
M SMT 2 CH2OH
M SBV 2 47 -0.7008 -0.0478
S SKP 5
ID FL5FAAGA0012
FORMULA C27H30O16
EXACTMASS 610.153384912
AVERAGEMASS 610.5175
SMILES c(c1)(O3)c(C(C(=C(c(c4)ccc(OC(O5)C(O)C(O)C(O)C5CO)c4)3)OC(C(O)2)OC(C(C(O)2)O)CO)=O)c(O)cc(O)1
M END