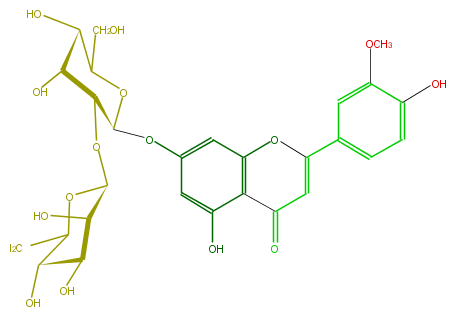
Mol:FL3FADGS0010
From Metabolomics.JP
(Difference between revisions)
| Line 1: | Line 1: | ||
| − | + | ||
| − | + | ||
| − | Copyright: ARM project http://www.metabolome.jp/ | + | Copyright: ARM project http://www.metabolome.jp/ |
| − | 44 48 0 0 0 0 0 0 0 0999 V2000 | + | 44 48 0 0 0 0 0 0 0 0999 V2000 |
| − | -0.9846 -0.3298 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.9846 -0.3298 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.9846 -1.0561 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.9846 -1.0561 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.3556 -1.4192 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.3556 -1.4192 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.2733 -1.0561 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.2733 -1.0561 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.2733 -0.3298 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.2733 -0.3298 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.3556 0.0333 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.3556 0.0333 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.9023 -1.4192 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.9023 -1.4192 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.5313 -1.0561 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.5313 -1.0561 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.5313 -0.3298 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.5313 -0.3298 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.9023 0.0333 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.9023 0.0333 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.9023 -2.1375 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.9023 -2.1375 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.1600 0.0332 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.1600 0.0332 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.8010 -0.3369 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.8010 -0.3369 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.4420 0.0332 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.4420 0.0332 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.4420 0.7734 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.4420 0.7734 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.8010 1.1436 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.8010 1.1436 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.1600 0.7734 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.1600 0.7734 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.3556 -2.1439 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.3556 -2.1439 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.1063 1.1569 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.1063 1.1569 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.6122 0.0327 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.6122 0.0327 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.0118 2.2223 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.0118 2.2223 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.3586 1.3995 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.3586 1.3995 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.7088 0.8780 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.7088 0.8780 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.3271 0.2331 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.3271 0.2331 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.1277 0.9769 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.1277 0.9769 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.7779 1.3860 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.7779 1.3860 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.8394 2.5488 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.8394 2.5488 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.6613 -0.0960 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.6613 -0.0960 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.8428 -2.4881 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.8428 -2.4881 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.9567 -2.3770 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.9567 -2.3770 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.8299 -1.5535 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.8299 -1.5535 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.4623 -0.9006 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.4623 -0.9006 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.2062 -1.0999 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.2062 -1.0999 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.2354 -1.8674 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.2354 -1.8674 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.0050 -3.2345 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.0050 -3.2345 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.3265 -2.9821 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.3265 -2.9821 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.7145 -1.4930 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.7145 -1.4930 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.8634 0.9983 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.8634 0.9983 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.6793 2.2377 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.6793 2.2377 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.3513 2.6285 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.3513 2.6285 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.8010 1.9495 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.8010 1.9495 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.3426 3.2345 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.3426 3.2345 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.1063 -2.1163 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.1063 -2.1163 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.5834 -3.0219 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.5834 -3.0219 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1 2 1 0 0 0 0 | + | 1 2 1 0 0 0 0 |
| − | 2 3 2 0 0 0 0 | + | 2 3 2 0 0 0 0 |
| − | 3 4 1 0 0 0 0 | + | 3 4 1 0 0 0 0 |
| − | 4 5 2 0 0 0 0 | + | 4 5 2 0 0 0 0 |
| − | 5 6 1 0 0 0 0 | + | 5 6 1 0 0 0 0 |
| − | 6 1 2 0 0 0 0 | + | 6 1 2 0 0 0 0 |
| − | 4 7 1 0 0 0 0 | + | 4 7 1 0 0 0 0 |
| − | 7 8 1 0 0 0 0 | + | 7 8 1 0 0 0 0 |
| − | 8 9 2 0 0 0 0 | + | 8 9 2 0 0 0 0 |
| − | 9 10 1 0 0 0 0 | + | 9 10 1 0 0 0 0 |
| − | 10 5 1 0 0 0 0 | + | 10 5 1 0 0 0 0 |
| − | 7 11 2 0 0 0 0 | + | 7 11 2 0 0 0 0 |
| − | 9 12 1 0 0 0 0 | + | 9 12 1 0 0 0 0 |
| − | 12 13 2 0 0 0 0 | + | 12 13 2 0 0 0 0 |
| − | 13 14 1 0 0 0 0 | + | 13 14 1 0 0 0 0 |
| − | 14 15 2 0 0 0 0 | + | 14 15 2 0 0 0 0 |
| − | 15 16 1 0 0 0 0 | + | 15 16 1 0 0 0 0 |
| − | 16 17 2 0 0 0 0 | + | 16 17 2 0 0 0 0 |
| − | 17 12 1 0 0 0 0 | + | 17 12 1 0 0 0 0 |
| − | 3 18 1 0 0 0 0 | + | 3 18 1 0 0 0 0 |
| − | 19 15 1 0 0 0 0 | + | 19 15 1 0 0 0 0 |
| − | 1 20 1 0 0 0 0 | + | 1 20 1 0 0 0 0 |
| − | 21 22 1 1 0 0 0 | + | 21 22 1 1 0 0 0 |
| − | 22 23 1 1 0 0 0 | + | 22 23 1 1 0 0 0 |
| − | 24 23 1 1 0 0 0 | + | 24 23 1 1 0 0 0 |
| − | 24 25 1 0 0 0 0 | + | 24 25 1 0 0 0 0 |
| − | 25 26 1 0 0 0 0 | + | 25 26 1 0 0 0 0 |
| − | 26 21 1 0 0 0 0 | + | 26 21 1 0 0 0 0 |
| − | 21 27 1 0 0 0 0 | + | 21 27 1 0 0 0 0 |
| − | 23 28 1 0 0 0 0 | + | 23 28 1 0 0 0 0 |
| − | 24 20 1 0 0 0 0 | + | 24 20 1 0 0 0 0 |
| − | 29 30 1 1 0 0 0 | + | 29 30 1 1 0 0 0 |
| − | 30 31 1 1 0 0 0 | + | 30 31 1 1 0 0 0 |
| − | 32 31 1 1 0 0 0 | + | 32 31 1 1 0 0 0 |
| − | 32 33 1 0 0 0 0 | + | 32 33 1 0 0 0 0 |
| − | 33 34 1 0 0 0 0 | + | 33 34 1 0 0 0 0 |
| − | 34 29 1 0 0 0 0 | + | 34 29 1 0 0 0 0 |
| − | 29 35 1 0 0 0 0 | + | 29 35 1 0 0 0 0 |
| − | 30 36 1 0 0 0 0 | + | 30 36 1 0 0 0 0 |
| − | 31 37 1 0 0 0 0 | + | 31 37 1 0 0 0 0 |
| − | 28 32 1 0 0 0 0 | + | 28 32 1 0 0 0 0 |
| − | 22 38 1 0 0 0 0 | + | 22 38 1 0 0 0 0 |
| − | 40 39 1 0 0 0 0 | + | 40 39 1 0 0 0 0 |
| − | 26 39 1 0 0 0 0 | + | 26 39 1 0 0 0 0 |
| − | 41 42 1 0 0 0 0 | + | 41 42 1 0 0 0 0 |
| − | 16 41 1 0 0 0 0 | + | 16 41 1 0 0 0 0 |
| − | 43 44 1 0 0 0 0 | + | 43 44 1 0 0 0 0 |
| − | 34 43 1 0 0 0 0 | + | 34 43 1 0 0 0 0 |
| − | M STY 1 1 SUP | + | M STY 1 1 SUP |
| − | M SLB 1 1 1 | + | M SLB 1 1 1 |
| − | M SAL 1 2 39 40 | + | M SAL 1 2 39 40 |
| − | M SBL 1 1 44 | + | M SBL 1 1 44 |
| − | M SMT 1 CH2OH | + | M SMT 1 CH2OH |
| − | M SBV 1 44 -0.0986 -0.8517 | + | M SBV 1 44 -0.0986 -0.8517 |
| − | M STY 1 2 SUP | + | M STY 1 2 SUP |
| − | M SLB 1 2 2 | + | M SLB 1 2 2 |
| − | M SAL 2 2 41 42 | + | M SAL 2 2 41 42 |
| − | M SBL 2 1 46 | + | M SBL 2 1 46 |
| − | M SMT 2 OCH3 | + | M SMT 2 OCH3 |
| − | M SBV 2 46 0.0000 -0.8060 | + | M SBV 2 46 0.0000 -0.8060 |
| − | M STY 1 3 SUP | + | M STY 1 3 SUP |
| − | M SLB 1 3 3 | + | M SLB 1 3 3 |
| − | M SAL 3 2 43 44 | + | M SAL 3 2 43 44 |
| − | M SBL 3 1 48 | + | M SBL 3 1 48 |
| − | M SMT 3 ^ CH2OH | + | M SMT 3 ^ CH2OH |
| − | M SBV 3 48 0.8709 0.2489 | + | M SBV 3 48 0.8709 0.2489 |
| − | S SKP 5 | + | S SKP 5 |
| − | ID FL3FADGS0010 | + | ID FL3FADGS0010 |
| − | FORMULA C28H32O16 | + | FORMULA C28H32O16 |
| − | EXACTMASS 624.1690349759999 | + | EXACTMASS 624.1690349759999 |
| − | AVERAGEMASS 624.54408 | + | AVERAGEMASS 624.54408 |
| − | SMILES c(c1)c(C(O5)=CC(=O)c(c54)c(cc(c4)OC(O3)C(C(C(C3CO)O)O)OC(C2O)OC(C(C2O)O)CO)O)cc(c1O)OC | + | SMILES c(c1)c(C(O5)=CC(=O)c(c54)c(cc(c4)OC(O3)C(C(C(C3CO)O)O)OC(C2O)OC(C(C2O)O)CO)O)cc(c1O)OC |
M END | M END | ||
| − | |||
Latest revision as of 09:00, 14 March 2009
Copyright: ARM project http://www.metabolome.jp/
44 48 0 0 0 0 0 0 0 0999 V2000
-0.9846 -0.3298 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.9846 -1.0561 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.3556 -1.4192 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.2733 -1.0561 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.2733 -0.3298 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.3556 0.0333 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.9023 -1.4192 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.5313 -1.0561 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.5313 -0.3298 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.9023 0.0333 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.9023 -2.1375 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.1600 0.0332 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.8010 -0.3369 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.4420 0.0332 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.4420 0.7734 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.8010 1.1436 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.1600 0.7734 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.3556 -2.1439 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
4.1063 1.1569 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.6122 0.0327 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.0118 2.2223 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.3586 1.3995 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.7088 0.8780 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.3271 0.2331 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.1277 0.9769 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.7779 1.3860 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.8394 2.5488 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.6613 -0.0960 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.8428 -2.4881 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.9567 -2.3770 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.8299 -1.5535 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.4623 -0.9006 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.2062 -1.0999 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.2354 -1.8674 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.0050 -3.2345 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.3265 -2.9821 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.7145 -1.4930 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.8634 0.9983 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.6793 2.2377 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.3513 2.6285 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.8010 1.9495 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.3426 3.2345 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.1063 -2.1163 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.5834 -3.0219 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1 2 1 0 0 0 0
2 3 2 0 0 0 0
3 4 1 0 0 0 0
4 5 2 0 0 0 0
5 6 1 0 0 0 0
6 1 2 0 0 0 0
4 7 1 0 0 0 0
7 8 1 0 0 0 0
8 9 2 0 0 0 0
9 10 1 0 0 0 0
10 5 1 0 0 0 0
7 11 2 0 0 0 0
9 12 1 0 0 0 0
12 13 2 0 0 0 0
13 14 1 0 0 0 0
14 15 2 0 0 0 0
15 16 1 0 0 0 0
16 17 2 0 0 0 0
17 12 1 0 0 0 0
3 18 1 0 0 0 0
19 15 1 0 0 0 0
1 20 1 0 0 0 0
21 22 1 1 0 0 0
22 23 1 1 0 0 0
24 23 1 1 0 0 0
24 25 1 0 0 0 0
25 26 1 0 0 0 0
26 21 1 0 0 0 0
21 27 1 0 0 0 0
23 28 1 0 0 0 0
24 20 1 0 0 0 0
29 30 1 1 0 0 0
30 31 1 1 0 0 0
32 31 1 1 0 0 0
32 33 1 0 0 0 0
33 34 1 0 0 0 0
34 29 1 0 0 0 0
29 35 1 0 0 0 0
30 36 1 0 0 0 0
31 37 1 0 0 0 0
28 32 1 0 0 0 0
22 38 1 0 0 0 0
40 39 1 0 0 0 0
26 39 1 0 0 0 0
41 42 1 0 0 0 0
16 41 1 0 0 0 0
43 44 1 0 0 0 0
34 43 1 0 0 0 0
M STY 1 1 SUP
M SLB 1 1 1
M SAL 1 2 39 40
M SBL 1 1 44
M SMT 1 CH2OH
M SBV 1 44 -0.0986 -0.8517
M STY 1 2 SUP
M SLB 1 2 2
M SAL 2 2 41 42
M SBL 2 1 46
M SMT 2 OCH3
M SBV 2 46 0.0000 -0.8060
M STY 1 3 SUP
M SLB 1 3 3
M SAL 3 2 43 44
M SBL 3 1 48
M SMT 3 ^ CH2OH
M SBV 3 48 0.8709 0.2489
S SKP 5
ID FL3FADGS0010
FORMULA C28H32O16
EXACTMASS 624.1690349759999
AVERAGEMASS 624.54408
SMILES c(c1)c(C(O5)=CC(=O)c(c54)c(cc(c4)OC(O3)C(C(C(C3CO)O)O)OC(C2O)OC(C(C2O)O)CO)O)cc(c1O)OC
M END