Mol:FL3FACGS0035
From Metabolomics.JP
(Difference between revisions)
| Line 1: | Line 1: | ||
| − | + | ||
| − | + | ||
| − | Copyright: ARM project http://www.metabolome.jp/ | + | Copyright: ARM project http://www.metabolome.jp/ |
| − | 43 47 0 0 0 0 0 0 0 0999 V2000 | + | 43 47 0 0 0 0 0 0 0 0999 V2000 |
| − | -3.7521 -0.8387 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.7521 -0.8387 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.7521 -1.6482 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.7521 -1.6482 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.0510 -2.0529 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.0510 -2.0529 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.3500 -1.6482 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.3500 -1.6482 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.3500 -0.8387 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.3500 -0.8387 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.0510 -0.4340 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.0510 -0.4340 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.6491 -2.0529 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.6491 -2.0529 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.9481 -1.6482 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.9481 -1.6482 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.9481 -0.8387 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.9481 -0.8387 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.6491 -0.4340 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.6491 -0.4340 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.6491 -2.8161 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.6491 -2.8161 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.2473 -0.4342 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.2473 -0.4342 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.4671 -0.8466 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.4671 -0.8466 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.1815 -0.4342 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.1815 -0.4342 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.1815 0.3907 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.1815 0.3907 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.4671 0.8033 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.4671 0.8033 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.2473 0.3907 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.2473 0.3907 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.5594 -0.3726 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.5594 -0.3726 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.0510 -2.8605 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.0510 -2.8605 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.8990 0.8118 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.8990 0.8118 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.4671 1.6281 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.4671 1.6281 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -5.0867 0.8059 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -5.0867 0.8059 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.4033 0.4113 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.4033 0.4113 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.6201 1.1701 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.6201 1.1701 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.4033 1.9334 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.4033 1.9334 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -5.0867 2.3281 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -5.0867 2.3281 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.8699 1.5692 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.8699 1.5692 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -5.6156 2.2995 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -5.6156 2.2995 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.5257 2.7132 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.5257 2.7132 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.0131 0.6517 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.0131 0.6517 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.0761 1.4287 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.0761 1.4287 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.6545 0.6982 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.6545 0.6982 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.4654 0.9300 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.4654 0.9300 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.2813 0.6982 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.2813 0.6982 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.7030 1.4287 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.7030 1.4287 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.8920 1.1970 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.8920 1.1970 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.2929 1.2187 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.2929 1.2187 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.4335 1.8358 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.4335 1.8358 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 5.3116 1.0773 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 5.3116 1.0773 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.8385 0.7272 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.8385 0.7272 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 5.6156 0.2785 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 5.6156 0.2785 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -5.3688 1.9185 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -5.3688 1.9185 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.8249 2.8605 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.8249 2.8605 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1 2 1 0 0 0 0 | + | 1 2 1 0 0 0 0 |
| − | 2 3 2 0 0 0 0 | + | 2 3 2 0 0 0 0 |
| − | 3 4 1 0 0 0 0 | + | 3 4 1 0 0 0 0 |
| − | 4 5 2 0 0 0 0 | + | 4 5 2 0 0 0 0 |
| − | 5 6 1 0 0 0 0 | + | 5 6 1 0 0 0 0 |
| − | 6 1 2 0 0 0 0 | + | 6 1 2 0 0 0 0 |
| − | 4 7 1 0 0 0 0 | + | 4 7 1 0 0 0 0 |
| − | 7 8 1 0 0 0 0 | + | 7 8 1 0 0 0 0 |
| − | 8 9 2 0 0 0 0 | + | 8 9 2 0 0 0 0 |
| − | 9 10 1 0 0 0 0 | + | 9 10 1 0 0 0 0 |
| − | 10 5 1 0 0 0 0 | + | 10 5 1 0 0 0 0 |
| − | 7 11 2 0 0 0 0 | + | 7 11 2 0 0 0 0 |
| − | 9 12 1 0 0 0 0 | + | 9 12 1 0 0 0 0 |
| − | 12 13 2 0 0 0 0 | + | 12 13 2 0 0 0 0 |
| − | 13 14 1 0 0 0 0 | + | 13 14 1 0 0 0 0 |
| − | 14 15 2 0 0 0 0 | + | 14 15 2 0 0 0 0 |
| − | 15 16 1 0 0 0 0 | + | 15 16 1 0 0 0 0 |
| − | 16 17 2 0 0 0 0 | + | 16 17 2 0 0 0 0 |
| − | 17 12 1 0 0 0 0 | + | 17 12 1 0 0 0 0 |
| − | 18 1 1 0 0 0 0 | + | 18 1 1 0 0 0 0 |
| − | 3 19 1 0 0 0 0 | + | 3 19 1 0 0 0 0 |
| − | 15 20 1 0 0 0 0 | + | 15 20 1 0 0 0 0 |
| − | 16 21 1 0 0 0 0 | + | 16 21 1 0 0 0 0 |
| − | 23 22 1 1 0 0 0 | + | 23 22 1 1 0 0 0 |
| − | 23 24 1 0 0 0 0 | + | 23 24 1 0 0 0 0 |
| − | 24 25 1 0 0 0 0 | + | 24 25 1 0 0 0 0 |
| − | 25 26 1 0 0 0 0 | + | 25 26 1 0 0 0 0 |
| − | 26 27 1 1 0 0 0 | + | 26 27 1 1 0 0 0 |
| − | 27 22 1 1 0 0 0 | + | 27 22 1 1 0 0 0 |
| − | 26 28 1 0 0 0 0 | + | 26 28 1 0 0 0 0 |
| − | 25 29 1 0 0 0 0 | + | 25 29 1 0 0 0 0 |
| − | 24 30 1 0 0 0 0 | + | 24 30 1 0 0 0 0 |
| − | 18 23 1 0 0 0 0 | + | 18 23 1 0 0 0 0 |
| − | 31 32 1 0 0 0 0 | + | 31 32 1 0 0 0 0 |
| − | 32 33 1 1 0 0 0 | + | 32 33 1 1 0 0 0 |
| − | 33 34 1 1 0 0 0 | + | 33 34 1 1 0 0 0 |
| − | 35 34 1 1 0 0 0 | + | 35 34 1 1 0 0 0 |
| − | 35 36 1 0 0 0 0 | + | 35 36 1 0 0 0 0 |
| − | 36 31 1 0 0 0 0 | + | 36 31 1 0 0 0 0 |
| − | 31 37 1 0 0 0 0 | + | 31 37 1 0 0 0 0 |
| − | 36 38 1 0 0 0 0 | + | 36 38 1 0 0 0 0 |
| − | 35 39 1 0 0 0 0 | + | 35 39 1 0 0 0 0 |
| − | 32 20 1 0 0 0 0 | + | 32 20 1 0 0 0 0 |
| − | 40 41 1 0 0 0 0 | + | 40 41 1 0 0 0 0 |
| − | 34 40 1 0 0 0 0 | + | 34 40 1 0 0 0 0 |
| − | 42 43 1 0 0 0 0 | + | 42 43 1 0 0 0 0 |
| − | 27 42 1 0 0 0 0 | + | 27 42 1 0 0 0 0 |
| − | M STY 1 1 SUP | + | M STY 1 1 SUP |
| − | M SLB 1 1 1 | + | M SLB 1 1 1 |
| − | M SAL 1 2 40 41 | + | M SAL 1 2 40 41 |
| − | M SBL 1 1 45 | + | M SBL 1 1 45 |
| − | M SMT 1 CH2OH | + | M SMT 1 CH2OH |
| − | M SBV 1 45 -0.5572 -0.0290 | + | M SBV 1 45 -0.5572 -0.0290 |
| − | M STY 1 2 SUP | + | M STY 1 2 SUP |
| − | M SLB 1 2 2 | + | M SLB 1 2 2 |
| − | M SAL 2 2 42 43 | + | M SAL 2 2 42 43 |
| − | M SBL 2 1 47 | + | M SBL 2 1 47 |
| − | M SMT 2 ^ CH2OH | + | M SMT 2 ^ CH2OH |
| − | M SBV 2 47 0.4989 -0.3492 | + | M SBV 2 47 0.4989 -0.3492 |
| − | S SKP 5 | + | S SKP 5 |
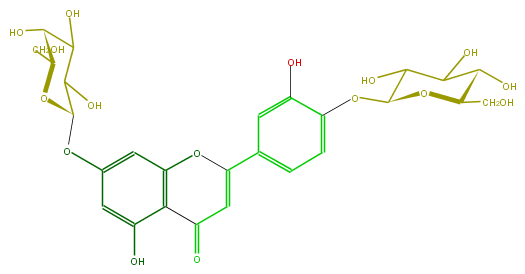
| − | ID FL3FACGS0035 | + | ID FL3FACGS0035 |
| − | FORMULA C27H30O16 | + | FORMULA C27H30O16 |
| − | EXACTMASS 610.153384912 | + | EXACTMASS 610.153384912 |
| − | AVERAGEMASS 610.5175 | + | AVERAGEMASS 610.5175 |
| − | SMILES c(O)(c4)c(c(cc(OC(C(O)5)OC(CO)C(O)C5O)4)1)C(=O)C=C(c(c2)cc(O)c(OC(O3)C(C(C(O)C(CO)3)O)O)c2)O1 | + | SMILES c(O)(c4)c(c(cc(OC(C(O)5)OC(CO)C(O)C5O)4)1)C(=O)C=C(c(c2)cc(O)c(OC(O3)C(C(C(O)C(CO)3)O)O)c2)O1 |
M END | M END | ||
| − | |||
Latest revision as of 09:00, 14 March 2009
Copyright: ARM project http://www.metabolome.jp/
43 47 0 0 0 0 0 0 0 0999 V2000
-3.7521 -0.8387 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.7521 -1.6482 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.0510 -2.0529 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.3500 -1.6482 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.3500 -0.8387 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.0510 -0.4340 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.6491 -2.0529 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.9481 -1.6482 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.9481 -0.8387 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.6491 -0.4340 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.6491 -2.8161 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.2473 -0.4342 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.4671 -0.8466 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.1815 -0.4342 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.1815 0.3907 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.4671 0.8033 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.2473 0.3907 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.5594 -0.3726 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.0510 -2.8605 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.8990 0.8118 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.4671 1.6281 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-5.0867 0.8059 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.4033 0.4113 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.6201 1.1701 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.4033 1.9334 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-5.0867 2.3281 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.8699 1.5692 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-5.6156 2.2995 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.5257 2.7132 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.0131 0.6517 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.0761 1.4287 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.6545 0.6982 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.4654 0.9300 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
4.2813 0.6982 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.7030 1.4287 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.8920 1.1970 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.2929 1.2187 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
4.4335 1.8358 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
5.3116 1.0773 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
4.8385 0.7272 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
5.6156 0.2785 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-5.3688 1.9185 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.8249 2.8605 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1 2 1 0 0 0 0
2 3 2 0 0 0 0
3 4 1 0 0 0 0
4 5 2 0 0 0 0
5 6 1 0 0 0 0
6 1 2 0 0 0 0
4 7 1 0 0 0 0
7 8 1 0 0 0 0
8 9 2 0 0 0 0
9 10 1 0 0 0 0
10 5 1 0 0 0 0
7 11 2 0 0 0 0
9 12 1 0 0 0 0
12 13 2 0 0 0 0
13 14 1 0 0 0 0
14 15 2 0 0 0 0
15 16 1 0 0 0 0
16 17 2 0 0 0 0
17 12 1 0 0 0 0
18 1 1 0 0 0 0
3 19 1 0 0 0 0
15 20 1 0 0 0 0
16 21 1 0 0 0 0
23 22 1 1 0 0 0
23 24 1 0 0 0 0
24 25 1 0 0 0 0
25 26 1 0 0 0 0
26 27 1 1 0 0 0
27 22 1 1 0 0 0
26 28 1 0 0 0 0
25 29 1 0 0 0 0
24 30 1 0 0 0 0
18 23 1 0 0 0 0
31 32 1 0 0 0 0
32 33 1 1 0 0 0
33 34 1 1 0 0 0
35 34 1 1 0 0 0
35 36 1 0 0 0 0
36 31 1 0 0 0 0
31 37 1 0 0 0 0
36 38 1 0 0 0 0
35 39 1 0 0 0 0
32 20 1 0 0 0 0
40 41 1 0 0 0 0
34 40 1 0 0 0 0
42 43 1 0 0 0 0
27 42 1 0 0 0 0
M STY 1 1 SUP
M SLB 1 1 1
M SAL 1 2 40 41
M SBL 1 1 45
M SMT 1 CH2OH
M SBV 1 45 -0.5572 -0.0290
M STY 1 2 SUP
M SLB 1 2 2
M SAL 2 2 42 43
M SBL 2 1 47
M SMT 2 ^ CH2OH
M SBV 2 47 0.4989 -0.3492
S SKP 5
ID FL3FACGS0035
FORMULA C27H30O16
EXACTMASS 610.153384912
AVERAGEMASS 610.5175
SMILES c(O)(c4)c(c(cc(OC(C(O)5)OC(CO)C(O)C5O)4)1)C(=O)C=C(c(c2)cc(O)c(OC(O3)C(C(C(O)C(CO)3)O)O)c2)O1
M END