Mol:FL3FAAGN0002
From Metabolomics.JP
(Difference between revisions)
| Line 1: | Line 1: | ||
| − | + | ||
| − | + | ||
| − | Copyright: ARM project http://www.metabolome.jp/ | + | Copyright: ARM project http://www.metabolome.jp/ |
| − | 43 47 0 0 0 0 0 0 0 0999 V2000 | + | 43 47 0 0 0 0 0 0 0 0999 V2000 |
| − | -1.4170 0.7365 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.4170 0.7365 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.4170 0.0941 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.4170 0.0941 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.8607 -0.2271 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.8607 -0.2271 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.3044 0.0941 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.3044 0.0941 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.3044 0.7365 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.3044 0.7365 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.8607 1.0577 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.8607 1.0577 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.2519 -0.2271 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.2519 -0.2271 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.8082 0.0941 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.8082 0.0941 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.8082 0.7365 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.8082 0.7365 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.2519 1.0577 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.2519 1.0577 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.2519 -0.7279 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.2519 -0.7279 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.3643 1.0575 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.3643 1.0575 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.9313 0.7302 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.9313 0.7302 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.4982 1.0575 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.4982 1.0575 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.4982 1.7122 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.4982 1.7122 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.9313 2.0396 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.9313 2.0396 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.3643 1.7122 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.3643 1.7122 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.9731 1.0575 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.9731 1.0575 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.0651 2.0395 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.0651 2.0395 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.8607 -0.7282 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.8607 -0.7282 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.5332 0.7398 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.5332 0.7398 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.0985 0.1660 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.0985 0.1660 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.4726 0.4094 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.4726 0.4094 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.7861 0.2922 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.7861 0.2922 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.3075 0.8549 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.3075 0.8549 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.9469 0.6253 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.9469 0.6253 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -5.4216 0.4065 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -5.4216 0.4065 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.7761 0.1330 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.7761 0.1330 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.1139 -0.1926 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.1139 -0.1926 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.7221 1.6601 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.7221 1.6601 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.3782 2.0390 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.3782 2.0390 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.7221 0.9025 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.7221 0.9025 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.3783 0.5237 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.3783 0.5237 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.3783 -0.2325 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.3783 -0.2325 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.8566 -0.5337 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.8566 -0.5337 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.8566 -1.1361 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.8566 -1.1361 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.3783 -1.4373 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.3783 -1.4373 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.9000 -1.1361 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.9000 -1.1361 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.9000 -0.5337 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.9000 -0.5337 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.3783 -2.0396 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.3783 -2.0396 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 5.4216 -1.4372 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 5.4216 -1.4372 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.9833 1.0238 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.9833 1.0238 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.7802 1.2373 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.7802 1.2373 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1 2 1 0 0 0 0 | + | 1 2 1 0 0 0 0 |
| − | 2 3 2 0 0 0 0 | + | 2 3 2 0 0 0 0 |
| − | 3 4 1 0 0 0 0 | + | 3 4 1 0 0 0 0 |
| − | 4 5 2 0 0 0 0 | + | 4 5 2 0 0 0 0 |
| − | 5 6 1 0 0 0 0 | + | 5 6 1 0 0 0 0 |
| − | 6 1 2 0 0 0 0 | + | 6 1 2 0 0 0 0 |
| − | 4 7 1 0 0 0 0 | + | 4 7 1 0 0 0 0 |
| − | 7 8 1 0 0 0 0 | + | 7 8 1 0 0 0 0 |
| − | 8 9 2 0 0 0 0 | + | 8 9 2 0 0 0 0 |
| − | 9 10 1 0 0 0 0 | + | 9 10 1 0 0 0 0 |
| − | 10 5 1 0 0 0 0 | + | 10 5 1 0 0 0 0 |
| − | 7 11 2 0 0 0 0 | + | 7 11 2 0 0 0 0 |
| − | 9 12 1 0 0 0 0 | + | 9 12 1 0 0 0 0 |
| − | 12 13 2 0 0 0 0 | + | 12 13 2 0 0 0 0 |
| − | 13 14 1 0 0 0 0 | + | 13 14 1 0 0 0 0 |
| − | 14 15 2 0 0 0 0 | + | 14 15 2 0 0 0 0 |
| − | 15 16 1 0 0 0 0 | + | 15 16 1 0 0 0 0 |
| − | 16 17 2 0 0 0 0 | + | 16 17 2 0 0 0 0 |
| − | 17 12 1 0 0 0 0 | + | 17 12 1 0 0 0 0 |
| − | 1 18 1 0 0 0 0 | + | 1 18 1 0 0 0 0 |
| − | 15 19 1 0 0 0 0 | + | 15 19 1 0 0 0 0 |
| − | 20 3 1 0 0 0 0 | + | 20 3 1 0 0 0 0 |
| − | 21 22 1 1 0 0 0 | + | 21 22 1 1 0 0 0 |
| − | 22 23 1 1 0 0 0 | + | 22 23 1 1 0 0 0 |
| − | 24 23 1 1 0 0 0 | + | 24 23 1 1 0 0 0 |
| − | 24 25 1 0 0 0 0 | + | 24 25 1 0 0 0 0 |
| − | 25 26 1 0 0 0 0 | + | 25 26 1 0 0 0 0 |
| − | 26 21 1 0 0 0 0 | + | 26 21 1 0 0 0 0 |
| − | 21 27 1 0 0 0 0 | + | 21 27 1 0 0 0 0 |
| − | 22 28 1 0 0 0 0 | + | 22 28 1 0 0 0 0 |
| − | 23 29 1 0 0 0 0 | + | 23 29 1 0 0 0 0 |
| − | 18 24 1 0 0 0 0 | + | 18 24 1 0 0 0 0 |
| − | 19 30 1 0 0 0 0 | + | 19 30 1 0 0 0 0 |
| − | 30 31 2 0 0 0 0 | + | 30 31 2 0 0 0 0 |
| − | 30 32 1 0 0 0 0 | + | 30 32 1 0 0 0 0 |
| − | 32 33 2 0 0 0 0 | + | 32 33 2 0 0 0 0 |
| − | 33 34 1 0 0 0 0 | + | 33 34 1 0 0 0 0 |
| − | 34 35 2 0 0 0 0 | + | 34 35 2 0 0 0 0 |
| − | 35 36 1 0 0 0 0 | + | 35 36 1 0 0 0 0 |
| − | 36 37 2 0 0 0 0 | + | 36 37 2 0 0 0 0 |
| − | 37 38 1 0 0 0 0 | + | 37 38 1 0 0 0 0 |
| − | 38 39 2 0 0 0 0 | + | 38 39 2 0 0 0 0 |
| − | 39 34 1 0 0 0 0 | + | 39 34 1 0 0 0 0 |
| − | 37 40 1 0 0 0 0 | + | 37 40 1 0 0 0 0 |
| − | 38 41 1 0 0 0 0 | + | 38 41 1 0 0 0 0 |
| − | 26 42 1 0 0 0 0 | + | 26 42 1 0 0 0 0 |
| − | 42 43 1 0 0 0 0 | + | 42 43 1 0 0 0 0 |
| − | M STY 1 1 SUP | + | M STY 1 1 SUP |
| − | M SLB 1 1 1 | + | M SLB 1 1 1 |
| − | M SAL 1 2 42 43 | + | M SAL 1 2 42 43 |
| − | M SBL 1 1 46 | + | M SBL 1 1 46 |
| − | M SMT 1 ^CH2OH | + | M SMT 1 ^CH2OH |
| − | M SBV 1 46 -6.5025 4.1049 | + | M SBV 1 46 -6.5025 4.1049 |
| − | S SKP 8 | + | S SKP 8 |
| − | ID FL3FAAGN0002 | + | ID FL3FAAGN0002 |
| − | KNApSAcK_ID C00004184 | + | KNApSAcK_ID C00004184 |
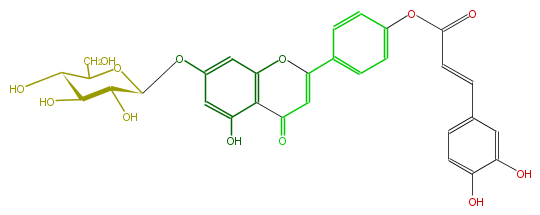
| − | NAME Apigenin 7-glucoside-4'-trans-caffeate | + | NAME Apigenin 7-glucoside-4'-trans-caffeate |
| − | CAS_RN 20196-92-3 | + | CAS_RN 20196-92-3 |
| − | FORMULA C30H26O13 | + | FORMULA C30H26O13 |
| − | EXACTMASS 594.137340918 | + | EXACTMASS 594.137340918 |
| − | AVERAGEMASS 594.51964 | + | AVERAGEMASS 594.51964 |
| − | SMILES C(Oc(c2)cc(O)c(C3=O)c(OC(c(c5)ccc(c5)OC(C=Cc(c4)ccc(O)c4O)=O)=C3)2)(C1O)OC(C(O)C1O)CO | + | SMILES C(Oc(c2)cc(O)c(C3=O)c(OC(c(c5)ccc(c5)OC(C=Cc(c4)ccc(O)c4O)=O)=C3)2)(C1O)OC(C(O)C1O)CO |
M END | M END | ||
| − | |||
Latest revision as of 09:00, 14 March 2009
Copyright: ARM project http://www.metabolome.jp/
43 47 0 0 0 0 0 0 0 0999 V2000
-1.4170 0.7365 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.4170 0.0941 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.8607 -0.2271 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.3044 0.0941 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.3044 0.7365 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.8607 1.0577 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.2519 -0.2271 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.8082 0.0941 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.8082 0.7365 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.2519 1.0577 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.2519 -0.7279 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.3643 1.0575 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.9313 0.7302 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.4982 1.0575 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.4982 1.7122 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.9313 2.0396 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.3643 1.7122 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.9731 1.0575 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.0651 2.0395 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.8607 -0.7282 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.5332 0.7398 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.0985 0.1660 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.4726 0.4094 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.7861 0.2922 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.3075 0.8549 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.9469 0.6253 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-5.4216 0.4065 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.7761 0.1330 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.1139 -0.1926 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.7221 1.6601 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.3782 2.0390 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.7221 0.9025 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.3783 0.5237 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.3783 -0.2325 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.8566 -0.5337 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.8566 -1.1361 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.3783 -1.4373 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.9000 -1.1361 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.9000 -0.5337 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.3783 -2.0396 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
5.4216 -1.4372 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.9833 1.0238 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.7802 1.2373 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1 2 1 0 0 0 0
2 3 2 0 0 0 0
3 4 1 0 0 0 0
4 5 2 0 0 0 0
5 6 1 0 0 0 0
6 1 2 0 0 0 0
4 7 1 0 0 0 0
7 8 1 0 0 0 0
8 9 2 0 0 0 0
9 10 1 0 0 0 0
10 5 1 0 0 0 0
7 11 2 0 0 0 0
9 12 1 0 0 0 0
12 13 2 0 0 0 0
13 14 1 0 0 0 0
14 15 2 0 0 0 0
15 16 1 0 0 0 0
16 17 2 0 0 0 0
17 12 1 0 0 0 0
1 18 1 0 0 0 0
15 19 1 0 0 0 0
20 3 1 0 0 0 0
21 22 1 1 0 0 0
22 23 1 1 0 0 0
24 23 1 1 0 0 0
24 25 1 0 0 0 0
25 26 1 0 0 0 0
26 21 1 0 0 0 0
21 27 1 0 0 0 0
22 28 1 0 0 0 0
23 29 1 0 0 0 0
18 24 1 0 0 0 0
19 30 1 0 0 0 0
30 31 2 0 0 0 0
30 32 1 0 0 0 0
32 33 2 0 0 0 0
33 34 1 0 0 0 0
34 35 2 0 0 0 0
35 36 1 0 0 0 0
36 37 2 0 0 0 0
37 38 1 0 0 0 0
38 39 2 0 0 0 0
39 34 1 0 0 0 0
37 40 1 0 0 0 0
38 41 1 0 0 0 0
26 42 1 0 0 0 0
42 43 1 0 0 0 0
M STY 1 1 SUP
M SLB 1 1 1
M SAL 1 2 42 43
M SBL 1 1 46
M SMT 1 ^CH2OH
M SBV 1 46 -6.5025 4.1049
S SKP 8
ID FL3FAAGN0002
KNApSAcK_ID C00004184
NAME Apigenin 7-glucoside-4'-trans-caffeate
CAS_RN 20196-92-3
FORMULA C30H26O13
EXACTMASS 594.137340918
AVERAGEMASS 594.51964
SMILES C(Oc(c2)cc(O)c(C3=O)c(OC(c(c5)ccc(c5)OC(C=Cc(c4)ccc(O)c4O)=O)=C3)2)(C1O)OC(C(O)C1O)CO
M END