Mol:FL1DA9NC0008
From Metabolomics.JP
(Difference between revisions)
| Line 1: | Line 1: | ||
| − | + | ||
| − | + | ||
| − | Copyright: ARM project http://www.metabolome.jp/ | + | Copyright: ARM project http://www.metabolome.jp/ |
| − | 36 38 0 0 0 0 0 0 0 0999 V2000 | + | 36 38 0 0 0 0 0 0 0 0999 V2000 |
| − | -0.8360 -0.5243 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.8360 -0.5243 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.8360 -1.1640 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.8360 -1.1640 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.2820 -1.4838 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.2820 -1.4838 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.2720 -1.1640 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.2720 -1.1640 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.2720 -0.5243 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.2720 -0.5243 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.2820 -0.2044 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.2820 -0.2044 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.8257 -1.4837 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.8257 -1.4837 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.3782 -1.1647 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.3782 -1.1647 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.9296 -1.4830 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.9296 -1.4830 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.4797 -1.1653 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.4797 -1.1653 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.0181 -1.4762 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.0181 -1.4762 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.5565 -1.1653 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.5565 -1.1653 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.5565 -0.5437 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.5565 -0.5437 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.0181 -0.2328 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.0181 -0.2328 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.4797 -0.5437 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.4797 -0.5437 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.8257 -2.1217 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.8257 -2.1217 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.2820 -2.1232 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.2820 -2.1232 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.2879 0.8580 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 | + | -0.2879 0.8580 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 |
| − | -0.8595 1.1880 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.8595 1.1880 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.8595 1.8115 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.8595 1.8115 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.3994 2.1232 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.3994 2.1232 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.9394 1.8115 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.9394 1.8115 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.9394 1.1880 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.9394 1.1880 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.3994 0.8763 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.3994 0.8763 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.8613 -0.1840 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.8613 -0.1840 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.8613 0.8059 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.8613 0.8059 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.2720 1.1462 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.2720 1.1462 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.2782 1.0466 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.2782 1.0466 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.4788 0.8766 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.4788 0.8766 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.0182 1.1880 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.0182 1.1880 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.4788 0.3809 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.4788 0.3809 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.5565 0.8773 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.5565 0.8773 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.3201 2.1229 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.3201 2.1229 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.1851 0.5343 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.1851 0.5343 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.3834 -0.9835 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.3834 -0.9835 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.0905 -1.6906 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.0905 -1.6906 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1 2 2 0 0 0 0 | + | 1 2 2 0 0 0 0 |
| − | 2 3 1 0 0 0 0 | + | 2 3 1 0 0 0 0 |
| − | 3 4 2 0 0 0 0 | + | 3 4 2 0 0 0 0 |
| − | 4 5 1 0 0 0 0 | + | 4 5 1 0 0 0 0 |
| − | 5 6 2 0 0 0 0 | + | 5 6 2 0 0 0 0 |
| − | 6 1 1 0 0 0 0 | + | 6 1 1 0 0 0 0 |
| − | 4 7 1 0 0 0 0 | + | 4 7 1 0 0 0 0 |
| − | 7 8 1 0 0 0 0 | + | 7 8 1 0 0 0 0 |
| − | 8 9 1 0 0 0 0 | + | 8 9 1 0 0 0 0 |
| − | 9 10 1 0 0 0 0 | + | 9 10 1 0 0 0 0 |
| − | 10 11 2 0 0 0 0 | + | 10 11 2 0 0 0 0 |
| − | 11 12 1 0 0 0 0 | + | 11 12 1 0 0 0 0 |
| − | 12 13 2 0 0 0 0 | + | 12 13 2 0 0 0 0 |
| − | 13 14 1 0 0 0 0 | + | 13 14 1 0 0 0 0 |
| − | 14 15 2 0 0 0 0 | + | 14 15 2 0 0 0 0 |
| − | 15 10 1 0 0 0 0 | + | 15 10 1 0 0 0 0 |
| − | 7 16 2 0 0 0 0 | + | 7 16 2 0 0 0 0 |
| − | 3 17 1 0 0 0 0 | + | 3 17 1 0 0 0 0 |
| − | 18 19 1 0 0 0 0 | + | 18 19 1 0 0 0 0 |
| − | 19 20 2 0 0 0 0 | + | 19 20 2 0 0 0 0 |
| − | 20 21 1 0 0 0 0 | + | 20 21 1 0 0 0 0 |
| − | 21 22 2 0 0 0 0 | + | 21 22 2 0 0 0 0 |
| − | 22 23 1 0 0 0 0 | + | 22 23 1 0 0 0 0 |
| − | 23 24 2 0 0 0 0 | + | 23 24 2 0 0 0 0 |
| − | 24 19 1 0 0 0 0 | + | 24 19 1 0 0 0 0 |
| − | 18 6 1 0 0 0 0 | + | 18 6 1 0 0 0 0 |
| − | 5 25 1 0 0 0 0 | + | 5 25 1 0 0 0 0 |
| − | 26 27 2 0 0 0 0 | + | 26 27 2 0 0 0 0 |
| − | 27 18 1 0 0 0 0 | + | 27 18 1 0 0 0 0 |
| − | 26 28 1 0 0 0 0 | + | 26 28 1 0 0 0 0 |
| − | 23 29 1 0 0 0 0 | + | 23 29 1 0 0 0 0 |
| − | 29 30 1 0 0 0 0 | + | 29 30 1 0 0 0 0 |
| − | 29 31 2 0 0 0 0 | + | 29 31 2 0 0 0 0 |
| − | 30 32 1 0 0 0 0 | + | 30 32 1 0 0 0 0 |
| − | 20 33 1 0 0 0 0 | + | 20 33 1 0 0 0 0 |
| − | 26 34 1 0 0 0 0 | + | 26 34 1 0 0 0 0 |
| − | 1 35 1 0 0 0 0 | + | 1 35 1 0 0 0 0 |
| − | 35 36 1 0 0 0 0 | + | 35 36 1 0 0 0 0 |
| − | M STY 1 2 SUP | + | M STY 1 2 SUP |
| − | M SLB 1 2 2 | + | M SLB 1 2 2 |
| − | M SAL 2 2 35 36 | + | M SAL 2 2 35 36 |
| − | M SBL 2 1 37 | + | M SBL 2 1 37 |
| − | M SMT 2 OCH3 | + | M SMT 2 OCH3 |
| − | M SVB 2 37 -1.1932 0.0945 | + | M SVB 2 37 -1.1932 0.0945 |
| − | M STY 1 1 SUP | + | M STY 1 1 SUP |
| − | M SLB 1 1 1 | + | M SLB 1 1 1 |
| − | M SAL 1 2 30 32 | + | M SAL 1 2 30 32 |
| − | M SBL 1 1 32 | + | M SBL 1 1 32 |
| − | M SMT 1 OCH3 | + | M SMT 1 OCH3 |
| − | M SVB 1 32 -3.0182 1.188 | + | M SVB 1 32 -3.0182 1.188 |
| − | S SKP 8 | + | S SKP 8 |
| − | ID FL1DA9NC0008 | + | ID FL1DA9NC0008 |
| − | KNApSAcK_ID C00007971 | + | KNApSAcK_ID C00007971 |
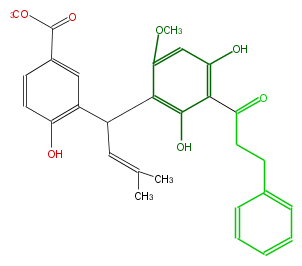
| − | NAME Piperaduncin A | + | NAME Piperaduncin A |
| − | CAS_RN 155023-54-4 | + | CAS_RN 155023-54-4 |
| − | FORMULA C29H30O7 | + | FORMULA C29H30O7 |
| − | EXACTMASS 490.199153314 | + | EXACTMASS 490.199153314 |
| − | AVERAGEMASS 490.5443 | + | AVERAGEMASS 490.5443 |
| − | SMILES Oc(c2)c(c(c(C(C=C(C)C)c(c3)c(O)ccc3C(=O)OC)c2OC)O)C(=O)CCc(c1)cccc1 | + | SMILES Oc(c2)c(c(c(C(C=C(C)C)c(c3)c(O)ccc3C(=O)OC)c2OC)O)C(=O)CCc(c1)cccc1 |
M END | M END | ||
| − | |||
Latest revision as of 09:00, 14 March 2009
Copyright: ARM project http://www.metabolome.jp/
36 38 0 0 0 0 0 0 0 0999 V2000
-0.8360 -0.5243 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.8360 -1.1640 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.2820 -1.4838 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.2720 -1.1640 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.2720 -0.5243 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.2820 -0.2044 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.8257 -1.4837 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.3782 -1.1647 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.9296 -1.4830 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.4797 -1.1653 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.0181 -1.4762 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.5565 -1.1653 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.5565 -0.5437 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.0181 -0.2328 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.4797 -0.5437 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.8257 -2.1217 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.2820 -2.1232 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.2879 0.8580 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0
-0.8595 1.1880 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.8595 1.8115 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.3994 2.1232 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.9394 1.8115 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.9394 1.1880 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.3994 0.8763 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.8613 -0.1840 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.8613 0.8059 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.2720 1.1462 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.2782 1.0466 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.4788 0.8766 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.0182 1.1880 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.4788 0.3809 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.5565 0.8773 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.3201 2.1229 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.1851 0.5343 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.3834 -0.9835 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.0905 -1.6906 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1 2 2 0 0 0 0
2 3 1 0 0 0 0
3 4 2 0 0 0 0
4 5 1 0 0 0 0
5 6 2 0 0 0 0
6 1 1 0 0 0 0
4 7 1 0 0 0 0
7 8 1 0 0 0 0
8 9 1 0 0 0 0
9 10 1 0 0 0 0
10 11 2 0 0 0 0
11 12 1 0 0 0 0
12 13 2 0 0 0 0
13 14 1 0 0 0 0
14 15 2 0 0 0 0
15 10 1 0 0 0 0
7 16 2 0 0 0 0
3 17 1 0 0 0 0
18 19 1 0 0 0 0
19 20 2 0 0 0 0
20 21 1 0 0 0 0
21 22 2 0 0 0 0
22 23 1 0 0 0 0
23 24 2 0 0 0 0
24 19 1 0 0 0 0
18 6 1 0 0 0 0
5 25 1 0 0 0 0
26 27 2 0 0 0 0
27 18 1 0 0 0 0
26 28 1 0 0 0 0
23 29 1 0 0 0 0
29 30 1 0 0 0 0
29 31 2 0 0 0 0
30 32 1 0 0 0 0
20 33 1 0 0 0 0
26 34 1 0 0 0 0
1 35 1 0 0 0 0
35 36 1 0 0 0 0
M STY 1 2 SUP
M SLB 1 2 2
M SAL 2 2 35 36
M SBL 2 1 37
M SMT 2 OCH3
M SVB 2 37 -1.1932 0.0945
M STY 1 1 SUP
M SLB 1 1 1
M SAL 1 2 30 32
M SBL 1 1 32
M SMT 1 OCH3
M SVB 1 32 -3.0182 1.188
S SKP 8
ID FL1DA9NC0008
KNApSAcK_ID C00007971
NAME Piperaduncin A
CAS_RN 155023-54-4
FORMULA C29H30O7
EXACTMASS 490.199153314
AVERAGEMASS 490.5443
SMILES Oc(c2)c(c(c(C(C=C(C)C)c(c3)c(O)ccc3C(=O)OC)c2OC)O)C(=O)CCc(c1)cccc1
M END