Mol:BMCCPPCL0003
From Metabolomics.JP
(Difference between revisions)
| Line 1: | Line 1: | ||
| − | + | ||
| − | + | ||
| − | Copyright: ARM project http://www.metabolome.jp/ | + | Copyright: ARM project http://www.metabolome.jp/ |
| − | 62 66 0 0 1 0 0 0 0 0999 V2000 | + | 62 66 0 0 1 0 0 0 0 0999 V2000 |
| − | 10.6281 -0.3402 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 10.6281 -0.3402 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 11.5499 -1.1275 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | 11.5499 -1.1275 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | 11.0860 -2.2475 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | 11.0860 -2.2475 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | 9.8774 -2.1524 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 9.8774 -2.1524 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 9.0459 -2.7080 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 9.0459 -2.7080 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 8.0651 -2.9031 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 8.0651 -2.9031 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 7.2778 -3.8249 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | 7.2778 -3.8249 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | 6.1578 -3.3610 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | 6.1578 -3.3610 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | 6.2529 -2.1524 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 6.2529 -2.1524 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 5.6973 -1.3210 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 5.6973 -1.3210 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 5.5022 -0.3402 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 5.5022 -0.3402 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.5804 0.4472 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.5804 0.4472 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 5.0443 1.5672 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 5.0443 1.5672 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 6.2529 1.4721 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 6.2529 1.4721 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 7.0844 2.0277 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 7.0844 2.0277 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 8.0651 2.2227 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 8.0651 2.2227 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 8.8525 3.1446 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 8.8525 3.1446 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 9.9725 2.6807 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 9.9725 2.6807 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 9.8774 1.4721 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 9.8774 1.4721 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 10.4330 0.6406 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 10.4330 0.6406 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 10.4330 -1.3210 0.0000 N 0 0 0 0 0 0 0 0 0 0 0 0 | + | 10.4330 -1.3210 0.0000 N 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 7.0844 -2.7080 0.0000 N 0 0 0 0 0 0 0 0 0 0 0 0 | + | 7.0844 -2.7080 0.0000 N 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 5.6973 0.6406 0.0000 N 0 0 0 0 0 0 0 0 0 0 0 0 | + | 5.6973 0.6406 0.0000 N 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 9.0459 2.0277 0.0000 N 0 0 0 0 0 0 0 0 0 0 0 0 | + | 9.0459 2.0277 0.0000 N 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 12.1994 -0.3671 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 12.1994 -0.3671 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 12.4738 -1.5102 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 12.4738 -1.5102 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 13.2671 -0.9014 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 13.2671 -0.9014 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 11.6085 -3.1002 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 11.6085 -3.1002 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 11.1313 -3.9790 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 11.1313 -3.9790 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 11.6538 -4.8316 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 11.6538 -4.8316 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 8.0382 -4.4744 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 8.0382 -4.4744 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 6.8951 -4.7488 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 6.8951 -4.7488 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 7.5039 -5.5422 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 7.5039 -5.5422 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 5.3051 -3.8835 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 5.3051 -3.8835 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 5.3313 -4.8832 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 5.3313 -4.8832 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.4787 -5.4057 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.4787 -5.4057 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.6080 0.2137 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.6080 0.2137 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.3240 -0.7451 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.3240 -0.7451 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.5218 2.4198 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.5218 2.4198 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.5222 2.3937 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.5222 2.3937 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.9997 3.2463 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.9997 3.2463 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 8.6190 4.1170 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 8.6190 4.1170 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 9.3444 4.8053 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 9.3444 4.8053 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 9.1110 5.7777 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 9.1110 5.7777 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 10.8252 3.2032 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 10.8252 3.2032 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 11.7040 2.7260 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 11.7040 2.7260 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 8.1521 6.0617 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 8.1521 6.0617 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 9.8363 6.4660 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 9.8363 6.4660 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 11.7302 1.7263 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 11.7302 1.7263 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 12.5566 3.2485 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 12.5566 3.2485 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.4768 4.1251 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.4768 4.1251 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.0000 3.2201 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.0000 3.2201 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.5999 -4.9285 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.5999 -4.9285 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.5048 -6.4053 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.5048 -6.4053 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.0123 -1.4705 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.0123 -1.4705 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.3516 -0.9785 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.3516 -0.9785 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 8.4953 -5.4116 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 8.4953 -5.4116 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 7.1212 -6.4660 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 7.1212 -6.4660 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 13.1366 0.0900 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 13.1366 0.0900 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 14.1910 -1.2841 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 14.1910 -1.2841 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 11.1767 -5.7105 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 11.1767 -5.7105 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 12.6535 -4.8055 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 12.6535 -4.8055 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 7 8 1 0 0 0 0 | + | 7 8 1 0 0 0 0 |
| − | 8 9 1 0 0 0 0 | + | 8 9 1 0 0 0 0 |
| − | 22 6 1 0 0 0 0 | + | 22 6 1 0 0 0 0 |
| − | 13 14 1 0 0 0 0 | + | 13 14 1 0 0 0 0 |
| − | 35 36 1 0 0 0 0 | + | 35 36 1 0 0 0 0 |
| − | 13 39 1 0 0 0 0 | + | 13 39 1 0 0 0 0 |
| − | 23 11 2 0 0 0 0 | + | 23 11 2 0 0 0 0 |
| − | 11 12 1 0 0 0 0 | + | 11 12 1 0 0 0 0 |
| − | 12 13 2 0 0 0 0 | + | 12 13 2 0 0 0 0 |
| − | 14 23 1 0 0 0 0 | + | 14 23 1 0 0 0 0 |
| − | 36 53 1 0 0 0 0 | + | 36 53 1 0 0 0 0 |
| − | 11 10 1 0 0 0 0 | + | 11 10 1 0 0 0 0 |
| − | 10 9 2 0 0 0 0 | + | 10 9 2 0 0 0 0 |
| − | 45 46 1 0 0 0 0 | + | 45 46 1 0 0 0 0 |
| − | 6 5 2 0 0 0 0 | + | 6 5 2 0 0 0 0 |
| − | 36 54 2 0 0 0 0 | + | 36 54 2 0 0 0 0 |
| − | 5 4 1 0 0 0 0 | + | 5 4 1 0 0 0 0 |
| − | 1 21 1 0 0 0 0 | + | 1 21 1 0 0 0 0 |
| − | 39 40 1 0 0 0 0 | + | 39 40 1 0 0 0 0 |
| − | 12 37 1 0 0 0 0 | + | 12 37 1 0 0 0 0 |
| − | 44 47 1 0 0 0 0 | + | 44 47 1 0 0 0 0 |
| − | 37 38 1 0 0 0 0 | + | 37 38 1 0 0 0 0 |
| − | 4 3 1 0 0 0 0 | + | 4 3 1 0 0 0 0 |
| − | 3 2 1 0 0 0 0 | + | 3 2 1 0 0 0 0 |
| − | 2 1 1 0 0 0 0 | + | 2 1 1 0 0 0 0 |
| − | 21 4 2 0 0 0 0 | + | 21 4 2 0 0 0 0 |
| − | 17 16 2 0 0 0 0 | + | 17 16 2 0 0 0 0 |
| − | 40 41 1 0 0 0 0 | + | 40 41 1 0 0 0 0 |
| − | 38 55 1 0 0 0 0 | + | 38 55 1 0 0 0 0 |
| − | 46 49 1 0 0 0 0 | + | 46 49 1 0 0 0 0 |
| − | 24 19 1 0 0 0 0 | + | 24 19 1 0 0 0 0 |
| − | 19 18 2 0 0 0 0 | + | 19 18 2 0 0 0 0 |
| − | 18 17 1 0 0 0 0 | + | 18 17 1 0 0 0 0 |
| − | 16 24 1 0 0 0 0 | + | 16 24 1 0 0 0 0 |
| − | 38 56 2 0 0 0 0 | + | 38 56 2 0 0 0 0 |
| − | 19 20 1 0 0 0 0 | + | 19 20 1 0 0 0 0 |
| − | 20 1 2 0 0 0 0 | + | 20 1 2 0 0 0 0 |
| − | 41 51 1 0 0 0 0 | + | 41 51 1 0 0 0 0 |
| − | 16 15 1 0 0 0 0 | + | 16 15 1 0 0 0 0 |
| − | 15 14 2 0 0 0 0 | + | 15 14 2 0 0 0 0 |
| − | 32 33 1 0 0 0 0 | + | 32 33 1 0 0 0 0 |
| − | 2 26 1 6 0 0 0 | + | 2 26 1 6 0 0 0 |
| − | 41 52 2 0 0 0 0 | + | 41 52 2 0 0 0 0 |
| − | 26 27 1 0 0 0 0 | + | 26 27 1 0 0 0 0 |
| − | 33 57 1 0 0 0 0 | + | 33 57 1 0 0 0 0 |
| − | 27 59 1 0 0 0 0 | + | 27 59 1 0 0 0 0 |
| − | 46 50 2 0 0 0 0 | + | 46 50 2 0 0 0 0 |
| − | 27 60 2 0 0 0 0 | + | 27 60 2 0 0 0 0 |
| − | 33 58 2 0 0 0 0 | + | 33 58 2 0 0 0 0 |
| − | 3 28 1 6 0 0 0 | + | 3 28 1 6 0 0 0 |
| − | 44 48 2 0 0 0 0 | + | 44 48 2 0 0 0 0 |
| − | 28 29 1 0 0 0 0 | + | 28 29 1 0 0 0 0 |
| − | 8 34 1 6 0 0 0 | + | 8 34 1 6 0 0 0 |
| − | 29 30 1 0 0 0 0 | + | 29 30 1 0 0 0 0 |
| − | 9 22 1 0 0 0 0 | + | 9 22 1 0 0 0 0 |
| − | 30 61 1 0 0 0 0 | + | 30 61 1 0 0 0 0 |
| − | 43 44 1 0 0 0 0 | + | 43 44 1 0 0 0 0 |
| − | 30 62 2 0 0 0 0 | + | 30 62 2 0 0 0 0 |
| − | 18 45 1 0 0 0 0 | + | 18 45 1 0 0 0 0 |
| − | 17 42 1 0 0 0 0 | + | 17 42 1 0 0 0 0 |
| − | 34 35 1 0 0 0 0 | + | 34 35 1 0 0 0 0 |
| − | 7 32 1 6 0 0 0 | + | 7 32 1 6 0 0 0 |
| − | 42 43 1 0 0 0 0 | + | 42 43 1 0 0 0 0 |
| − | 7 31 1 1 0 0 0 | + | 7 31 1 1 0 0 0 |
| − | 6 7 1 0 0 0 0 | + | 6 7 1 0 0 0 0 |
| − | 2 25 1 1 0 0 0 | + | 2 25 1 1 0 0 0 |
| − | S SKP 7 | + | S SKP 7 |
| − | ID BMCCPPCL0003 | + | ID BMCCPPCL0003 |
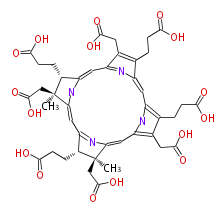
| − | NAME Sirohydrochlorin | + | NAME Sirohydrochlorin |
| − | FORMULA C42H46N4O16 | + | FORMULA C42H46N4O16 |
| − | EXACTMASS 862.2908 | + | EXACTMASS 862.2908 |
| − | AVERAGEMASS 862.832 | + | AVERAGEMASS 862.832 |
| − | SMILES c(c2)([C@H]5CCC(O)=O)nc([C@](CC(O)=O)(C)5)cc([C@@H](CCC(O)=O)1)nc(cc(c4CC(O)=O)nc(c4CCC(O)=O)cc(c(CCC(O)=O)3)nc2c3CC(O)=O)[C@@](CC(O)=O)1C | + | SMILES c(c2)([C@H]5CCC(O)=O)nc([C@](CC(O)=O)(C)5)cc([C@@H](CCC(O)=O)1)nc(cc(c4CC(O)=O)nc(c4CCC(O)=O)cc(c(CCC(O)=O)3)nc2c3CC(O)=O)[C@@](CC(O)=O)1C |
| − | KEGG http://www.genome.jp/dbget-bin/www_bget?compound+C05778 | + | KEGG http://www.genome.jp/dbget-bin/www_bget?compound+C05778 |
M END | M END | ||
| − | |||
Latest revision as of 09:00, 14 March 2009
Copyright: ARM project http://www.metabolome.jp/
62 66 0 0 1 0 0 0 0 0999 V2000
10.6281 -0.3402 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
11.5499 -1.1275 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
11.0860 -2.2475 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
9.8774 -2.1524 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
9.0459 -2.7080 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
8.0651 -2.9031 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
7.2778 -3.8249 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
6.1578 -3.3610 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
6.2529 -2.1524 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
5.6973 -1.3210 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
5.5022 -0.3402 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.5804 0.4472 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
5.0443 1.5672 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
6.2529 1.4721 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
7.0844 2.0277 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
8.0651 2.2227 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
8.8525 3.1446 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
9.9725 2.6807 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
9.8774 1.4721 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
10.4330 0.6406 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
10.4330 -1.3210 0.0000 N 0 0 0 0 0 0 0 0 0 0 0 0
7.0844 -2.7080 0.0000 N 0 0 0 0 0 0 0 0 0 0 0 0
5.6973 0.6406 0.0000 N 0 0 0 0 0 0 0 0 0 0 0 0
9.0459 2.0277 0.0000 N 0 0 0 0 0 0 0 0 0 0 0 0
12.1994 -0.3671 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
12.4738 -1.5102 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
13.2671 -0.9014 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
11.6085 -3.1002 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
11.1313 -3.9790 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
11.6538 -4.8316 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
8.0382 -4.4744 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
6.8951 -4.7488 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
7.5039 -5.5422 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
5.3051 -3.8835 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
5.3313 -4.8832 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.4787 -5.4057 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.6080 0.2137 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.3240 -0.7451 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.5218 2.4198 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.5222 2.3937 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.9997 3.2463 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
8.6190 4.1170 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
9.3444 4.8053 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
9.1110 5.7777 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
10.8252 3.2032 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
11.7040 2.7260 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
8.1521 6.0617 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
9.8363 6.4660 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
11.7302 1.7263 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
12.5566 3.2485 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.4768 4.1251 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.0000 3.2201 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.5999 -4.9285 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
4.5048 -6.4053 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
4.0123 -1.4705 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.3516 -0.9785 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
8.4953 -5.4116 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
7.1212 -6.4660 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
13.1366 0.0900 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
14.1910 -1.2841 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
11.1767 -5.7105 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
12.6535 -4.8055 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
7 8 1 0 0 0 0
8 9 1 0 0 0 0
22 6 1 0 0 0 0
13 14 1 0 0 0 0
35 36 1 0 0 0 0
13 39 1 0 0 0 0
23 11 2 0 0 0 0
11 12 1 0 0 0 0
12 13 2 0 0 0 0
14 23 1 0 0 0 0
36 53 1 0 0 0 0
11 10 1 0 0 0 0
10 9 2 0 0 0 0
45 46 1 0 0 0 0
6 5 2 0 0 0 0
36 54 2 0 0 0 0
5 4 1 0 0 0 0
1 21 1 0 0 0 0
39 40 1 0 0 0 0
12 37 1 0 0 0 0
44 47 1 0 0 0 0
37 38 1 0 0 0 0
4 3 1 0 0 0 0
3 2 1 0 0 0 0
2 1 1 0 0 0 0
21 4 2 0 0 0 0
17 16 2 0 0 0 0
40 41 1 0 0 0 0
38 55 1 0 0 0 0
46 49 1 0 0 0 0
24 19 1 0 0 0 0
19 18 2 0 0 0 0
18 17 1 0 0 0 0
16 24 1 0 0 0 0
38 56 2 0 0 0 0
19 20 1 0 0 0 0
20 1 2 0 0 0 0
41 51 1 0 0 0 0
16 15 1 0 0 0 0
15 14 2 0 0 0 0
32 33 1 0 0 0 0
2 26 1 6 0 0 0
41 52 2 0 0 0 0
26 27 1 0 0 0 0
33 57 1 0 0 0 0
27 59 1 0 0 0 0
46 50 2 0 0 0 0
27 60 2 0 0 0 0
33 58 2 0 0 0 0
3 28 1 6 0 0 0
44 48 2 0 0 0 0
28 29 1 0 0 0 0
8 34 1 6 0 0 0
29 30 1 0 0 0 0
9 22 1 0 0 0 0
30 61 1 0 0 0 0
43 44 1 0 0 0 0
30 62 2 0 0 0 0
18 45 1 0 0 0 0
17 42 1 0 0 0 0
34 35 1 0 0 0 0
7 32 1 6 0 0 0
42 43 1 0 0 0 0
7 31 1 1 0 0 0
6 7 1 0 0 0 0
2 25 1 1 0 0 0
S SKP 7
ID BMCCPPCL0003
NAME Sirohydrochlorin
FORMULA C42H46N4O16
EXACTMASS 862.2908
AVERAGEMASS 862.832
SMILES c(c2)([C@H]5CCC(O)=O)nc([C@](CC(O)=O)(C)5)cc([C@@H](CCC(O)=O)1)nc(cc(c4CC(O)=O)nc(c4CCC(O)=O)cc(c(CCC(O)=O)3)nc2c3CC(O)=O)[C@@](CC(O)=O)1C
KEGG http://www.genome.jp/dbget-bin/www_bget?compound+C05778
M END