Mol:FL5FAGGS0010
From Metabolomics.JP
(Difference between revisions)
| Line 1: | Line 1: | ||
| − | + | ||
| − | + | ||
| − | Copyright: ARM project http://www.metabolome.jp/ | + | Copyright: ARM project http://www.metabolome.jp/ |
| − | 44 48 0 0 0 0 0 0 0 0999 V2000 | + | 44 48 0 0 0 0 0 0 0 0999 V2000 |
| − | -0.8252 0.5551 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.8252 0.5551 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.8252 -0.2700 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.8252 -0.2700 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.1107 -0.6826 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.1107 -0.6826 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.6038 -0.2700 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.6038 -0.2700 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.6038 0.5551 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.6038 0.5551 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.1107 0.9675 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.1107 0.9675 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.3183 -0.6826 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.3183 -0.6826 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.0328 -0.2700 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.0328 -0.2700 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.0328 0.5551 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.0328 0.5551 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.3183 0.9675 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.3183 0.9675 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.3183 -1.3258 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.3183 -1.3258 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.9326 1.1264 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.9326 1.1264 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.6608 0.7059 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.6608 0.7059 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.3890 1.1264 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.3890 1.1264 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.3890 1.9673 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.3890 1.9673 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.6608 2.3877 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.6608 2.3877 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.9326 1.9673 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.9326 1.9673 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.1107 -1.5073 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.1107 -1.5073 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 5.1683 2.4172 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 5.1683 2.4172 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.6787 1.0478 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.6787 1.0478 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.8726 -0.7798 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.8726 -0.7798 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.6608 3.2283 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.6608 3.2283 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 5.1170 0.7060 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 5.1170 0.7060 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.6375 -2.1369 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.6375 -2.1369 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.3252 -2.5339 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.3252 -2.5339 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.1070 -1.7703 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.1070 -1.7703 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.3252 -1.0023 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.3252 -1.0023 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.6375 -0.6051 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.6375 -0.6051 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.8556 -1.3687 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.8556 -1.3687 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.8318 -1.3791 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.8318 -1.3791 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.6228 -2.8456 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.6228 -2.8456 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.0667 -3.2283 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.0667 -3.2283 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.6128 -2.2268 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.6128 -2.2268 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.1277 1.0459 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.1277 1.0459 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.6511 0.4167 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.6511 0.4167 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.9645 0.6837 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.9645 0.6837 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.3021 0.6908 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.3021 0.6908 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.7834 1.1723 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.7834 1.1723 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.3840 0.8553 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.3840 0.8553 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.7304 0.8053 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.7304 0.8053 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.3884 0.4005 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.3884 0.4005 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.3223 0.2694 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.3223 0.2694 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.9399 1.4312 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.9399 1.4312 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -5.1683 1.8064 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -5.1683 1.8064 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1 2 1 0 0 0 0 | + | 1 2 1 0 0 0 0 |
| − | 2 3 2 0 0 0 0 | + | 2 3 2 0 0 0 0 |
| − | 4 5 2 0 0 0 0 | + | 4 5 2 0 0 0 0 |
| − | 5 6 1 0 0 0 0 | + | 5 6 1 0 0 0 0 |
| − | 6 1 2 0 0 0 0 | + | 6 1 2 0 0 0 0 |
| − | 4 7 1 0 0 0 0 | + | 4 7 1 0 0 0 0 |
| − | 7 8 1 0 0 0 0 | + | 7 8 1 0 0 0 0 |
| − | 8 9 2 0 0 0 0 | + | 8 9 2 0 0 0 0 |
| − | 9 10 1 0 0 0 0 | + | 9 10 1 0 0 0 0 |
| − | 10 5 1 0 0 0 0 | + | 10 5 1 0 0 0 0 |
| − | 7 11 2 0 0 0 0 | + | 7 11 2 0 0 0 0 |
| − | 9 12 1 0 0 0 0 | + | 9 12 1 0 0 0 0 |
| − | 12 13 2 0 0 0 0 | + | 12 13 2 0 0 0 0 |
| − | 13 14 1 0 0 0 0 | + | 13 14 1 0 0 0 0 |
| − | 14 15 2 0 0 0 0 | + | 14 15 2 0 0 0 0 |
| − | 15 16 1 0 0 0 0 | + | 15 16 1 0 0 0 0 |
| − | 16 17 2 0 0 0 0 | + | 16 17 2 0 0 0 0 |
| − | 17 12 1 0 0 0 0 | + | 17 12 1 0 0 0 0 |
| − | 3 18 1 0 0 0 0 | + | 3 18 1 0 0 0 0 |
| − | 19 15 1 0 0 0 0 | + | 19 15 1 0 0 0 0 |
| − | 4 3 1 0 0 0 0 | + | 4 3 1 0 0 0 0 |
| − | 1 20 1 0 0 0 0 | + | 1 20 1 0 0 0 0 |
| − | 21 8 1 0 0 0 0 | + | 21 8 1 0 0 0 0 |
| − | 16 22 1 0 0 0 0 | + | 16 22 1 0 0 0 0 |
| − | 14 23 1 0 0 0 0 | + | 14 23 1 0 0 0 0 |
| − | 25 24 1 1 0 0 0 | + | 25 24 1 1 0 0 0 |
| − | 25 26 1 0 0 0 0 | + | 25 26 1 0 0 0 0 |
| − | 26 27 1 0 0 0 0 | + | 26 27 1 0 0 0 0 |
| − | 27 28 1 0 0 0 0 | + | 27 28 1 0 0 0 0 |
| − | 28 29 1 1 0 0 0 | + | 28 29 1 1 0 0 0 |
| − | 29 24 1 1 0 0 0 | + | 29 24 1 1 0 0 0 |
| − | 29 30 1 0 0 0 0 | + | 29 30 1 0 0 0 0 |
| − | 24 31 1 0 0 0 0 | + | 24 31 1 0 0 0 0 |
| − | 25 32 1 0 0 0 0 | + | 25 32 1 0 0 0 0 |
| − | 26 33 1 0 0 0 0 | + | 26 33 1 0 0 0 0 |
| − | 28 21 1 0 0 0 0 | + | 28 21 1 0 0 0 0 |
| − | 34 35 1 1 0 0 0 | + | 34 35 1 1 0 0 0 |
| − | 35 36 1 1 0 0 0 | + | 35 36 1 1 0 0 0 |
| − | 37 36 1 1 0 0 0 | + | 37 36 1 1 0 0 0 |
| − | 37 38 1 0 0 0 0 | + | 37 38 1 0 0 0 0 |
| − | 38 39 1 0 0 0 0 | + | 38 39 1 0 0 0 0 |
| − | 39 34 1 0 0 0 0 | + | 39 34 1 0 0 0 0 |
| − | 34 40 1 0 0 0 0 | + | 34 40 1 0 0 0 0 |
| − | 35 41 1 0 0 0 0 | + | 35 41 1 0 0 0 0 |
| − | 36 42 1 0 0 0 0 | + | 36 42 1 0 0 0 0 |
| − | 37 20 1 0 0 0 0 | + | 37 20 1 0 0 0 0 |
| − | 43 44 1 0 0 0 0 | + | 43 44 1 0 0 0 0 |
| − | 39 43 1 0 0 0 0 | + | 39 43 1 0 0 0 0 |
| − | M STY 1 1 SUP | + | M STY 1 1 SUP |
| − | M SLB 1 1 1 | + | M SLB 1 1 1 |
| − | M SAL 1 2 43 44 | + | M SAL 1 2 43 44 |
| − | M SBL 1 1 48 | + | M SBL 1 1 48 |
| − | M SMT 1 ^ CH2OH | + | M SMT 1 ^ CH2OH |
| − | M SBV 1 48 0.5559 -0.5758 | + | M SBV 1 48 0.5559 -0.5758 |
| − | S SKP 5 | + | S SKP 5 |
| − | ID FL5FAGGS0010 | + | ID FL5FAGGS0010 |
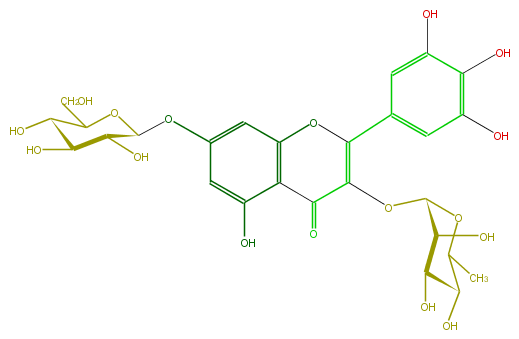
| − | FORMULA C27H30O17 | + | FORMULA C27H30O17 |
| − | EXACTMASS 626.148299534 | + | EXACTMASS 626.148299534 |
| − | AVERAGEMASS 626.5169000000001 | + | AVERAGEMASS 626.5169000000001 |
| − | SMILES C(C(O)5)(OC(CO)C(O)C5O)Oc(c1)cc(O3)c(C(=O)C(=C3c(c4)cc(c(c(O)4)O)O)OC(C(O)2)OC(C)C(C(O)2)O)c1O | + | SMILES C(C(O)5)(OC(CO)C(O)C5O)Oc(c1)cc(O3)c(C(=O)C(=C3c(c4)cc(c(c(O)4)O)O)OC(C(O)2)OC(C)C(C(O)2)O)c1O |
M END | M END | ||
| − | |||
Latest revision as of 09:00, 14 March 2009
Copyright: ARM project http://www.metabolome.jp/
44 48 0 0 0 0 0 0 0 0999 V2000
-0.8252 0.5551 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.8252 -0.2700 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.1107 -0.6826 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.6038 -0.2700 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.6038 0.5551 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.1107 0.9675 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.3183 -0.6826 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.0328 -0.2700 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.0328 0.5551 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.3183 0.9675 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.3183 -1.3258 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.9326 1.1264 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.6608 0.7059 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.3890 1.1264 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.3890 1.9673 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.6608 2.3877 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.9326 1.9673 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.1107 -1.5073 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
5.1683 2.4172 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.6787 1.0478 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.8726 -0.7798 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.6608 3.2283 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
5.1170 0.7060 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.6375 -2.1369 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.3252 -2.5339 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.1070 -1.7703 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.3252 -1.0023 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.6375 -0.6051 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.8556 -1.3687 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.8318 -1.3791 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.6228 -2.8456 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
4.0667 -3.2283 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
4.6128 -2.2268 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.1277 1.0459 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.6511 0.4167 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.9645 0.6837 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.3021 0.6908 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.7834 1.1723 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.3840 0.8553 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.7304 0.8053 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.3884 0.4005 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.3223 0.2694 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.9399 1.4312 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-5.1683 1.8064 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1 2 1 0 0 0 0
2 3 2 0 0 0 0
4 5 2 0 0 0 0
5 6 1 0 0 0 0
6 1 2 0 0 0 0
4 7 1 0 0 0 0
7 8 1 0 0 0 0
8 9 2 0 0 0 0
9 10 1 0 0 0 0
10 5 1 0 0 0 0
7 11 2 0 0 0 0
9 12 1 0 0 0 0
12 13 2 0 0 0 0
13 14 1 0 0 0 0
14 15 2 0 0 0 0
15 16 1 0 0 0 0
16 17 2 0 0 0 0
17 12 1 0 0 0 0
3 18 1 0 0 0 0
19 15 1 0 0 0 0
4 3 1 0 0 0 0
1 20 1 0 0 0 0
21 8 1 0 0 0 0
16 22 1 0 0 0 0
14 23 1 0 0 0 0
25 24 1 1 0 0 0
25 26 1 0 0 0 0
26 27 1 0 0 0 0
27 28 1 0 0 0 0
28 29 1 1 0 0 0
29 24 1 1 0 0 0
29 30 1 0 0 0 0
24 31 1 0 0 0 0
25 32 1 0 0 0 0
26 33 1 0 0 0 0
28 21 1 0 0 0 0
34 35 1 1 0 0 0
35 36 1 1 0 0 0
37 36 1 1 0 0 0
37 38 1 0 0 0 0
38 39 1 0 0 0 0
39 34 1 0 0 0 0
34 40 1 0 0 0 0
35 41 1 0 0 0 0
36 42 1 0 0 0 0
37 20 1 0 0 0 0
43 44 1 0 0 0 0
39 43 1 0 0 0 0
M STY 1 1 SUP
M SLB 1 1 1
M SAL 1 2 43 44
M SBL 1 1 48
M SMT 1 ^ CH2OH
M SBV 1 48 0.5559 -0.5758
S SKP 5
ID FL5FAGGS0010
FORMULA C27H30O17
EXACTMASS 626.148299534
AVERAGEMASS 626.5169000000001
SMILES C(C(O)5)(OC(CO)C(O)C5O)Oc(c1)cc(O3)c(C(=O)C(=C3c(c4)cc(c(c(O)4)O)O)OC(C(O)2)OC(C)C(C(O)2)O)c1O
M END